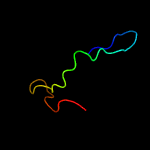
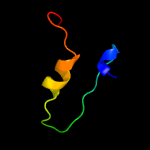
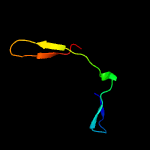
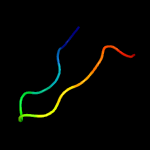
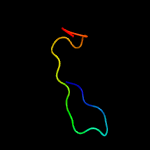
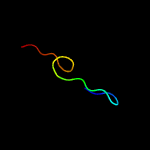
1 c2m0oA_
30.5
41
PDB header: peptide binding proteinChain: A: PDB Molecule: phd finger protein 1;PDBTitle: the solution structure of human phf1 in complex with h3k36me3
2 d1iu4a_
28.3
38
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Microbial transglutaminase
3 c3iu0A_
27.1
38
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: structural basis for zymogen activation and substrate binding of2 transglutaminase from streptomyces mobaraense
4 c1rfoC_
25.7
40
PDB header: viral proteinChain: C: PDB Molecule: whisker antigen control protein;PDBTitle: trimeric foldon of the t4 phagehead fibritin
5 c3co5B_
24.7
28
PDB header: transcription regulatorChain: B: PDB Molecule: putative two-component system transcriptional responsePDBTitle: crystal structure of sigma-54 interaction domain of putative2 transcriptional response regulator from neisseria gonorrhoeae
6 c6g49A_
24.3
33
PDB header: transferaseChain: A: PDB Molecule: protein-glutamine gamma-glutamyltransferase;PDBTitle: crystal structure of the periplasmic domain of tgpa from pseudomonas2 aeruginosa
7 c2lp7C_
19.4
40
PDB header: viral proteinChain: C: PDB Molecule: envelope glycoprotein;PDBTitle: structure of gp41-m-mat, a membrane associated mper trimer from hiv-12 gp41.
8 c6dkuA_
18.9
40
PDB header: unknown functionChain: A: PDB Molecule: vp35;PDBTitle: crystal structure of myotis vp35 mutant of interferon inhibitory2 domain
9 c3isrB_
18.6
29
PDB header: hydrolaseChain: B: PDB Molecule: transglutaminase-like enzymes, putative cysteine protease;PDBTitle: the crystal structure of a putative cysteine protease from cytophaga2 hutchinsonii to 1.9a
10 c4gh9A_
16.9
40
PDB header: viral protein,rna binding proteinChain: A: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of marburg virus vp35 rna binding domain
11 c4xhiB_
15.7
33
PDB header: transcriptionChain: B: PDB Molecule: rna-dependent rna polymerase;PDBTitle: crystal structure of native thosea asigna virus rna-dependent rna2 polymerase (rdrp) at 2.15 angstrom resolution
12 c2m5tA_
15.5
50
PDB header: viral proteinChain: A: PDB Molecule: human rhinovirus 2a proteinase;PDBTitle: solution structure of the 2a proteinase from a common cold agent,2 human rhinovirus rv-c02, strain w12
13 c6d9oA_
15.2
70
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant e25a
14 c1avyA_
14.6
40
PDB header: coiled coilChain: A: PDB Molecule: fibritin;PDBTitle: fibritin deletion mutant m (bacteriophage t4)
15 c3fkeB_
14.4
53
PDB header: rna binding proteinChain: B: PDB Molecule: polymerase cofactor vp35;PDBTitle: structure of the ebola vp35 interferon inhibitory domain
16 c3ks8D_
14.0
47
PDB header: viral protein/rnaChain: D: PDB Molecule: polymerase cofactor vp35;PDBTitle: crystal structure of reston ebolavirus vp35 rna binding domain in2 complex with 18bp dsrna
17 c1ox3A_
13.8
40
PDB header: chaperoneChain: A: PDB Molecule: fibritin;PDBTitle: crystal structure of mini-fibritin
18 c1z8rA_
13.6
60
PDB header: hydrolaseChain: A: PDB Molecule: coxsackievirus b4 polyprotein;PDBTitle: 2a cysteine proteinase from human coxsackievirus b4 (strain2 jvb / benschoten / new york / 51)
19 c4p78D_
13.5
14
PDB header: toxinChain: D: PDB Molecule: hica3 toxin;PDBTitle: hica3 and hicb3 toxin-antitoxin complex
20 c3rp7A_
13.4
56
PDB header: oxidoreductaseChain: A: PDB Molecule: flavoprotein monooxygenase;PDBTitle: crystal structure of klebsiella pneumoniae hpxo complexed with fad and2 uric acid
21 c1v1hB_
not modelled
13.3
39
PDB header: adenovirusChain: B: PDB Molecule: fibritin, fiber protein;PDBTitle: adenovirus fibre shaft sequence n-terminally fused to the2 bacteriophage t4 fibritin foldon trimerisation motif with a short3 linker
22 c5fn0C_
not modelled
12.9
38
PDB header: oxidoreductaseChain: C: PDB Molecule: kynurenine 3-monooxygenase;PDBTitle: crystal structure of pseudomonas fluorescens kynurenine-3-2 monooxygenase (kmo) in complex with gsk180
23 c3j21B_
not modelled
12.5
19
PDB header: ribosomeChain: B: PDB Molecule: 50s ribosomal protein l2p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
24 d2hrva_
not modelled
12.4
40
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral cysteine protease of trypsin fold
25 c3c4aA_
not modelled
12.0
31
PDB header: oxidoreductaseChain: A: PDB Molecule: probable tryptophan hydroxylase viod;PDBTitle: crystal structure of viod hydroxylase in complex with fad from2 chromobacterium violaceum. northeast structural genomics consortium3 target cvr158
26 c2l1sA_
not modelled
11.8
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yohn;PDBTitle: yp_001336205.1
27 c2dkhA_
not modelled
11.6
33
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-hydroxybenzoate hydroxylase;PDBTitle: crystal structure of 3-hydroxybenzoate hydroxylase from comamonas2 testosteroni, in complex with the substrate
28 c3gmbB_
not modelled
10.1
39
PDB header: oxidoreductaseChain: B: PDB Molecule: 2-methyl-3-hydroxypyridine-5-carboxylic acid oxygenase;PDBTitle: crystal structure of 2-methyl-3-hydroxypyridine-5-carboxylic acid2 oxygenase
29 c1vw4d_
not modelled
10.0
86
PDB header: ribosomeChain: D: PDB Molecule: PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
30 c2qa1A_
not modelled
10.0
39
PDB header: oxidoreductaseChain: A: PDB Molecule: polyketide oxygenase pgae;PDBTitle: crystal structure of pgae, an aromatic hydroxylase involved in2 angucycline biosynthesis
31 c1pn0A_
not modelled
9.9
38
PDB header: oxidoreductaseChain: A: PDB Molecule: phenol 2-monooxygenase;PDBTitle: phenol hydroxylase from trichosporon cutaneum
32 d1pn0a1
not modelled
9.9
38
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD-linked reductases, N-terminal domain
33 c6h3jA_
not modelled
9.5
32
PDB header: protein transportChain: A: PDB Molecule: protein involved in gliding motility spra;PDBTitle: structural snapshots of the type 9 protein translocon plug-complex
34 d1m1ha1
not modelled
9.3
64
Fold: N-utilization substance G protein NusG, insert domainSuperfamily: N-utilization substance G protein NusG, insert domainFamily: N-utilization substance G protein NusG, insert domain
35 c2a56A_
not modelled
9.2
27
PDB header: luminescent proteinChain: A: PDB Molecule: gfp-like non-fluorescent chromoprotein fp595 chain 1;PDBTitle: fluorescent protein asfp595, a143s, on-state, 5min irradiation
36 c1phhA_
not modelled
9.2
39
PDB header: oxidoreductaseChain: A: PDB Molecule: p-hydroxybenzoate hydroxylase;PDBTitle: crystal structure of p-hydroxybenzoate hydroxylase complexed with its2 reaction product 3,4-dihydroxybenzoate
37 d1jmab2
not modelled
9.0
55
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
38 c3kzsD_
not modelled
8.8
21
PDB header: hydrolaseChain: D: PDB Molecule: glycosyl hydrolase family 5;PDBTitle: crystal structure of glycosyl hydrolase family 5 (np_809925.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
39 c6h3iA_
not modelled
8.7
32
PDB header: protein transportChain: A: PDB Molecule: protein involved in gliding motility spra;PDBTitle: structural snapshots of the type 9 protein translocon
40 c3lf4A_
not modelled
8.7
33
PDB header: fluorescent proteinChain: A: PDB Molecule: fluorescent timer precursor blue102;PDBTitle: crystal structure of fluorescent timer precursor blue102
41 c4c26A_
not modelled
8.6
29
PDB header: toxinChain: A: PDB Molecule: hica;PDBTitle: solution nmr structure of the hica toxin from burkholderia2 pseudomallei
42 c2e6iA_
not modelled
8.5
32
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase itk/tsk;PDBTitle: solution structure of the btk motif of tyrosine-protein2 kinase itk from human
43 c2l62A_
not modelled
8.4
58
PDB header: metal binding proteinChain: A: PDB Molecule: ec protein i/ii;PDBTitle: protein and metal cluster structure of the wheat metallothionein2 domain g-ec-1. the second part of the puzzle.
44 c4yl5A_
not modelled
8.0
40
PDB header: transferaseChain: A: PDB Molecule: putative phosphomethylpyrimidine kinase;PDBTitle: structure of a putative phosphomethylpyrimidine kinase from2 acinetobacter baumannii
45 c3es4B_
not modelled
7.8
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf861 with a rmlc-like cupin fold;PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
46 c3e1tA_
not modelled
7.7
28
PDB header: flavoproteinChain: A: PDB Molecule: halogenase;PDBTitle: structure and action of the myxobacterial chondrochloren2 halogenase cndh, a new variant of fad-dependent halogenases
47 c4k22A_
not modelled
7.7
33
PDB header: oxidoreductaseChain: A: PDB Molecule: protein visc;PDBTitle: structure of the c-terminal truncated form of e.coli c5-hydroxylase2 ubii involved in ubiquinone (q8) biosynthesis
48 c2gw4C_
not modelled
7.6
40
PDB header: luminescent proteinChain: C: PDB Molecule: kaede;PDBTitle: crystal structure of stony coral fluorescent protein kaede, red form
49 c2m2rA_
not modelled
7.5
75
PDB header: unknown functionChain: A: PDB Molecule: inhibitor cystine knot peptide mch-2;PDBTitle: solution structure of mch-2: a novel inhibitor cystine knot peptide2 from momordica charantia
50 c2l61A_
not modelled
7.4
58
PDB header: metal binding proteinChain: A: PDB Molecule: ec protein i/ii;PDBTitle: protein and metal cluster structure of the wheat metallothionein2 domain g-ec-1. the second part of the puzzle.
51 d1k8kb1
not modelled
7.2
22
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
52 c3i3lA_
not modelled
7.1
33
PDB header: hydrolaseChain: A: PDB Molecule: alkylhalidase cmls;PDBTitle: crystal structure of cmls, a flavin-dependent halogenase
53 c2mfpA_
not modelled
7.0
43
PDB header: metal binding proteinChain: A: PDB Molecule: ec protein i/ii;PDBTitle: solution structure of the circular g-domain analog from the wheat2 metallothionein ec-1
54 c2vouA_
not modelled
7.0
50
PDB header: oxidoreductaseChain: A: PDB Molecule: 2,6-dihydroxypyridine hydroxylase;PDBTitle: structure of 2,6-dihydroxypyridine-3-hydroxylase from2 arthrobacter nicotinovorans
55 c6fviA_
not modelled
7.0
58
PDB header: cell cycleChain: A: PDB Molecule: centrosomal protein of 192 kda;PDBTitle: ash / papd-like domain of human cep192 (papd-like domain 7)
56 d1d0gr2
not modelled
6.9
42
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
57 c6d93A_
not modelled
6.9
60
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant y31a
58 c2m2qA_
not modelled
6.9
75
PDB header: unknown functionChain: A: PDB Molecule: inhibitor cystine knot peptide mch-1;PDBTitle: solution structure of mch-1: a novel inhibitor cystine knot peptide2 from momordica charantia
59 c3fmwC_
not modelled
6.7
44
PDB header: oxidoreductaseChain: C: PDB Molecule: oxygenase;PDBTitle: the crystal structure of mtmoiv, a baeyer-villiger monooxygenase from2 the mithramycin biosynthetic pathway in streptomyces argillaceus.
60 c4xa9g_
not modelled
6.7
62
PDB header: structural genomics, unknown functionChain: G: PDB Molecule: gala protein type 1, 3 or 4;PDBTitle: crystal structure of the complex between the n-terminal domain of ravj2 and legl1 from legionella pneumophila str. philadelphia
61 c5evyX_
not modelled
6.7
50
PDB header: oxidoreductaseChain: X: PDB Molecule: salicylate hydroxylase;PDBTitle: salicylate hydroxylase substrate complex
62 d2nn6c2
not modelled
6.6
40
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like
63 c2r0gB_
not modelled
6.5
39
PDB header: oxidoreductaseChain: B: PDB Molecule: rebc;PDBTitle: chromopyrrolic acid-soaked rebc with bound 7-carboxy-k252c
64 d1bkta_
not modelled
6.5
100
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
65 c5kowA_
not modelled
6.4
39
PDB header: oxidoreductaseChain: A: PDB Molecule: pentachlorophenol 4-monooxygenase;PDBTitle: structure of rifampicin monooxygenase
66 c2rgjA_
not modelled
6.4
39
PDB header: oxidoreductaseChain: A: PDB Molecule: flavin-containing monooxygenase;PDBTitle: crystal structure of flavin-containing monooxygenase phzs
67 c2xdoC_
not modelled
6.2
28
PDB header: oxidoreductaseChain: C: PDB Molecule: tetx2 protein;PDBTitle: structure of the tetracycline degrading monooxygenase tetx2 from2 bacteroides thetaiotaomicron
68 c4hb9A_
not modelled
5.9
33
PDB header: oxidoreductaseChain: A: PDB Molecule: similarities with probable monooxygenase;PDBTitle: crystal structure of a putative fad containing monooxygenase from2 photorhabdus luminescens subsp. laumondii tto1 (target psi-012791)
69 c5c0rA_
not modelled
5.8
40
PDB header: viral protein/immune systemChain: A: PDB Molecule: hemagglutinin, envelope glycoprotein, fibritin fusionPDBTitle: crystal structure of a generation 3 influenza hemagglutinin stabilized2 stem complexed with the broadly neutralizing antibody c179
70 d1g2ra_
not modelled
5.8
44
Fold: YlxR-likeSuperfamily: YlxR-likeFamily: YlxR-like
71 c4n9xA_
not modelled
5.7
39
PDB header: oxidoreductaseChain: A: PDB Molecule: putative monooxygenase;PDBTitle: crystal structure of the octaprenyl-methyl-methoxy-benzq molecule from2 erwina carotovora subsp. atroseptica strain scri 1043 / atcc baa-672,3 northeast structural genomics consortium (nesg) target ewr161
72 c6c6rA_
not modelled
5.7
28
PDB header: oxidoreductase/oxidoreductase inhibitorChain: A: PDB Molecule: squalene monooxygenase;PDBTitle: human squalene epoxidase (sqle, squalene monooxygenase) structure with2 fad
73 c6d8tA_
not modelled
5.7
60
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant e25k/k27e
74 c4bk2A_
not modelled
5.6
28
PDB header: oxidoreductaseChain: A: PDB Molecule: probable salicylate monooxygenase;PDBTitle: crystal structure of 3-hydroxybenzoate 6-hydroxylase2 uncovers lipid-assisted flavoprotein strategy for3 regioselective aromatic hydroxylation: q301e mutant
75 c6d8sA_
not modelled
5.5
60
PDB header: toxinChain: A: PDB Molecule: potassium channel toxin alpha-ktx 5.4;PDBTitle: nmr solution structure of tamapin, mutant k27e
76 c4ebuA_
not modelled
5.4
26
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxygluconokinase;PDBTitle: crystal structure of a sugar kinase (target efi-502312) from2 oceanicola granulosus, with bound amp/adp crystal form i
77 c6aioA_
not modelled
5.4
44
PDB header: flavoproteinChain: A: PDB Molecule: pnpa;PDBTitle: crystal structure of p-nitrophenol 4-monooxygenase pnpa from2 pseudomonas putida dll-e4
78 c5dbjA_
not modelled
5.3
33
PDB header: flavoproteinChain: A: PDB Molecule: fadh2-dependent halogenase plta;PDBTitle: crystal structure of halogenase plta
79 c3nvnA_
not modelled
5.3
71
PDB header: viral protein/signaling proteinChain: A: PDB Molecule: evm139;PDBTitle: molecular mechanism of guidance cue recognition
80 d2ktxa_
not modelled
5.2
80
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: Scorpion toxin-likeFamily: Short-chain scorpion toxins
81 c4k2xB_
not modelled
5.1
39
PDB header: oxidoreductase, flavoproteinChain: B: PDB Molecule: polyketide oxygenase/hydroxylase;PDBTitle: oxys anhydrotetracycline hydroxylase from streptomyces rimosus
82 c3ihgA_
not modelled
5.1
39
PDB header: flavoprotein, oxidoreductaseChain: A: PDB Molecule: rdme;PDBTitle: crystal structure of a ternary complex of aklavinone-11 hydroxylase2 with fad and aklavinone
83 c1dmfA_
not modelled
5.1
71
PDB header: metallothioneinChain: A: PDB Molecule: cd6 metallothionein-1;PDBTitle: the three-dimensional solution structure of callinectes2 sapidus metallothionein-i determined by homonuclear and3 heteronuclear magnetic resonance spectoscopy