1 c6ic4C_
99.9
30
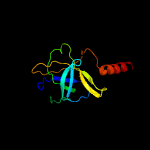
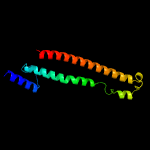
PDB header: protein transportChain: C: PDB Molecule: toluene tolerance efflux transporter (abc superfamily,PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
2 c5uw8C_
99.8
20
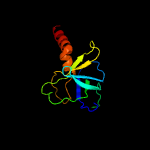
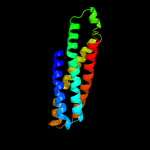
PDB header: transport proteinChain: C: PDB Molecule: probable phospholipid abc transporter-binding protein mlad;PDBTitle: structure of e. coli mce protein mlad, core mce domain
3 c5uvnD_
99.6
23
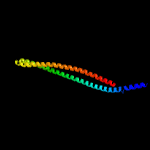
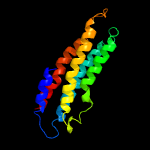
PDB header: transport proteinChain: D: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
4 c5uvnA_
99.6
23
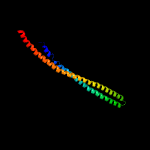
PDB header: transport proteinChain: A: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
5 c5uvnC_
99.6
23
PDB header: transport proteinChain: C: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
6 c5uvnE_
99.6
23
PDB header: transport proteinChain: E: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
7 c5uvnF_
99.6
23
PDB header: transport proteinChain: F: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
8 c5uvnB_
99.6
23
PDB header: transport proteinChain: B: PDB Molecule: paraquat-inducible protein b;PDBTitle: structure of e. coli mce protein pqib, periplasmic domain
9 c3g67A_
95.2
7
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of a soluble chemoreceptor from thermotoga2 maritima
10 c1qu7A_
93.9
8
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: four helical-bundle structure of the cytoplasmic domain of a serine2 chemotaxis receptor
11 c3vkhA_
88.8
7
PDB header: motor proteinChain: A: PDB Molecule: dynein heavy chain, cytoplasmic;PDBTitle: x-ray structure of a functional full-length dynein motor domain
12 c6b7nC_
87.0
10
PDB header: viral proteinChain: C: PDB Molecule: spike protein;PDBTitle: cryo-electron microscopy structure of porcine delta coronavirus spike2 protein in the pre-fusion state
13 c2ieqC_
86.3
11
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: core structure of s2 from the human coronavirus nl63 spike2 glycoprotein
14 c6e6aB_
85.3
17
PDB header: protein bindingChain: B: PDB Molecule: inclusion membrane protein a;PDBTitle: triclinic crystal form of inca g144a point mutant
15 c3lnrA_
84.7
18
PDB header: signaling proteinChain: A: PDB Molecule: aerotaxis transducer aer2;PDBTitle: crystal structure of poly-hamp domains from the p. aeruginosa soluble2 receptor aer2
16 c3zx6A_
83.9
11
PDB header: signalingChain: A: PDB Molecule: hamp, methyl-accepting chemotaxis protein i;PDBTitle: structure of hamp(af1503)-tsr fusion - hamp (a291v) mutant
17 d1eq1a_
83.7
11
Fold: Apolipophorin-IIISuperfamily: Apolipophorin-IIIFamily: Apolipophorin-III
18 c5u0pU_
83.6
11
PDB header: transcriptionChain: U: PDB Molecule: mediator complex subunit 21;PDBTitle: cryo-em structure of the transcriptional mediator
19 c2ch7A_
82.5
12
PDB header: chemotaxisChain: A: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: crystal structure of the cytoplasmic domain of a bacterial2 chemoreceptor from thermotoga maritima
20 c2j5uB_
79.5
12
PDB header: cell shape regulationChain: B: PDB Molecule: mrec protein;PDBTitle: mrec lysteria monocytogenes
21 c2d4yA_
not modelled
79.4
13
PDB header: structural proteinChain: A: PDB Molecule: flagellar hook-associated protein 1;PDBTitle: crystal structure of a 49k fragment of hap1 (flgk)
22 c6ezvX_
not modelled
77.5
11
PDB header: toxinChain: X: PDB Molecule: non-hemolytic enterotoxin lytic component l1;PDBTitle: the cytotoxin maka from vibrio cholerae
23 d1st6a4
not modelled
77.0
14
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin
24 c1deqO_
not modelled
73.1
9
PDB header: blood clottingChain: O: PDB Molecule: fibrinogen (beta chain);PDBTitle: the crystal structure of modified bovine fibrinogen (at ~42 angstrom resolution)
25 c6gajA_
not modelled
72.7
9
PDB header: viral proteinChain: A: PDB Molecule: outer capsid protein sigma-1;PDBTitle: crystal structure of the t1l reovirus sigma1 coiled coil tail (iodide)
26 c2wpqA_
not modelled
72.5
5
PDB header: membrane proteinChain: A: PDB Molecule: trimeric autotransporter adhesin fragment;PDBTitle: salmonella enterica sada 479-519 fused to gcn4 adaptors (sadak3, in-2 register fusion)
27 c3ojaB_
not modelled
71.5
13
PDB header: protein bindingChain: B: PDB Molecule: anopheles plasmodium-responsive leucine-rich repeat proteinPDBTitle: crystal structure of lrim1/apl1c complex
28 c6nb3B_
not modelled
70.2
17
PDB header: virusChain: B: PDB Molecule: spike glycoprotein;PDBTitle: mers-cov complex with human neutralizing lca60 antibody fab fragment2 (state 1)
29 c1deqF_
not modelled
69.4
13
PDB header: blood clottingChain: F: PDB Molecule: fibrinogen (gamma chain);PDBTitle: the crystal structure of modified bovine fibrinogen (at ~42 angstrom resolution)
30 c1ei3E_
not modelled
68.2
14
PDB header: blood clottingChain: E: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
31 c4rh7A_
not modelled
68.1
9
PDB header: motor proteinChain: A: PDB Molecule: green fluorescent protein/cytoplasmic dynein 2 heavy chainPDBTitle: crystal structure of human cytoplasmic dynein 2 motor domain in2 complex with adp.vi
32 c5zhyA_
not modelled
65.7
13
PDB header: viral proteinChain: A: PDB Molecule: spike glycoprotein, spike glycoprotein;PDBTitle: structural characterization of the hcov-229e fusion core
33 c5xbjA_
not modelled
61.8
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: flagellar hook-associated protein flgk;PDBTitle: the structure of the flagellar hook junction protein hap1 (flgk) from2 campylobacter jejuni
34 c4ut1A_
not modelled
61.7
18
PDB header: motor proteinChain: A: PDB Molecule: flagellar hook-associated protein;PDBTitle: the structure of the flagellar hook junction protein flgk2 from burkholderia pseudomallei
35 c2qihA_
not modelled
60.7
11
PDB header: cell adhesionChain: A: PDB Molecule: protein uspa1;PDBTitle: crystal structure of 527-665 fragment of uspa1 protein from moraxella2 catarrhalis
36 c6o7ua_
not modelled
59.4
11
PDB header: membrane proteinChain: A: PDB Molecule: PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
37 d1g4us1
not modelled
58.6
12
Fold: Four-helical up-and-down bundleSuperfamily: Bacterial GAP domainFamily: Bacterial GAP domain
38 c3cwgA_
not modelled
57.1
11
PDB header: transcriptionChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: unphosphorylated mouse stat3 core fragment
39 c6gapB_
not modelled
54.5
13
PDB header: viral proteinChain: B: PDB Molecule: outer capsid protein sigma-1;PDBTitle: crystal structure of the t3d reovirus sigma1 coiled coil tail and body
40 c1kmiZ_
not modelled
53.8
10
PDB header: signaling proteinChain: Z: PDB Molecule: chemotaxis protein chez;PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
41 c2kbbA_
not modelled
49.8
13
PDB header: structural proteinChain: A: PDB Molecule: talin-1;PDBTitle: nmr structure of the talin rod domain, 1655-1822
42 c6gaoC_
not modelled
47.0
9
PDB header: viral proteinChain: C: PDB Molecule: outer capsid protein sigma-1;PDBTitle: crystal structure of the t1l reovirus sigma1 coiled coil tail and body
43 d1st6a3
not modelled
45.1
14
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin
44 c2p22A_
not modelled
44.6
12
PDB header: transport proteinChain: A: PDB Molecule: suppressor protein stp22 of temperature-sensitive alpha-PDBTitle: structure of the yeast escrt-i heterotetramer core
45 c5n76C_
not modelled
43.8
15
PDB header: nickel-binding proteinChain: C: PDB Molecule: coot;PDBTitle: crystal structure of the apo-form of the co dehydrogenase accessory2 protein coot from rhodospirillum rubrum
46 c2qf4A_
not modelled
43.7
14
PDB header: structural proteinChain: A: PDB Molecule: cell shape determining protein mrec;PDBTitle: high resolution structure of the major periplasmic domain from the2 cell shape-determining filament mrec (orthorhombic form)
47 c2yfaA_
not modelled
43.3
10
PDB header: receptorChain: A: PDB Molecule: methyl-accepting chemotaxis transducer;PDBTitle: x-ray structure of mcps ligand binding domain in complex with malate
48 c2dq3A_
not modelled
42.9
10
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
49 c5cwsJ_
not modelled
42.7
10
PDB header: protein transportChain: J: PDB Molecule: nucleoporin nup49;PDBTitle: crystal structure of the intact chaetomium thermophilum nsp1-nup49-2 nup57 channel nucleoporin heterotrimer bound to its nic96 nuclear3 pore complex attachment site
50 d1ykhb1
not modelled
42.4
11
Fold: Mediator hinge subcomplex-likeSuperfamily: Mediator hinge subcomplex-likeFamily: CSE2-like
51 d1h9ma2
not modelled
41.4
23
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain
52 c5j65A_
not modelled
41.2
9
PDB header: toxinChain: A: PDB Molecule: pesticidal crystal protein cry6aa;PDBTitle: crystal structure of trypsin activated cry6aa
53 c5zuvB_
not modelled
40.4
4
PDB header: viral protein, inhibitorChain: B: PDB Molecule: spike glycoprotein,spike glycoprotein,inhibitor ek1;PDBTitle: crystal structure of the human coronavirus 229e hr1 motif in complex2 with pan-covs inhibitor ek1
54 c3j6vL_
not modelled
40.0
18
PDB header: ribosomeChain: L: PDB Molecule: 28s ribosomal protein s12, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
55 d1st6a5
not modelled
39.7
16
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin
56 c3ur1C_
not modelled
39.3
9
PDB header: immune systemChain: C: PDB Molecule: methyl-accepting chemotaxis protein;PDBTitle: the structure of a ternary complex between chea domains p4 and p5 with2 chew and with a truncated fragment of tm14, a chemoreceptor analog3 from thermotoga maritima.
57 d1quua1
not modelled
39.1
11
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat
58 c2vs0B_
not modelled
39.1
13
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
59 c3ghgI_
not modelled
38.3
14
PDB header: blood clottingChain: I: PDB Molecule: fibrinogen gamma chain;PDBTitle: crystal structure of human fibrinogen
60 c4lwsA_
not modelled
37.6
12
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
61 c4njlA_
not modelled
37.3
10
PDB header: viral proteinChain: A: PDB Molecule: s protein;PDBTitle: crystal structure of middle east respiratory syndrome coronavirus s22 protein fusion core
62 c1ei3C_
not modelled
37.2
14
PDB header: blood clottingChain: C: PDB Molecule: fibrinogen;PDBTitle: crystal structure of native chicken fibrinogen
63 d2uubl1
not modelled
36.9
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
64 d1h9ra2
not modelled
36.5
20
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain
65 c1bf5A_
not modelled
34.9
8
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcription 1-PDBTitle: tyrosine phosphorylated stat-1/dna complex
66 d1h9ma1
not modelled
34.5
23
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain
67 d1guta_
not modelled
34.4
16
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP
68 c2nrjA_
not modelled
34.2
11
PDB header: toxinChain: A: PDB Molecule: hbl b protein;PDBTitle: crystal structure of hemolysin binding component from2 bacillus cereus
69 c6grjG_
not modelled
34.1
10
PDB header: toxinChain: G: PDB Molecule: ahlb;PDBTitle: structure of the ahlb pore of the tripartite alpha-pore forming toxin,2 ahl, from aeromonas hydrophila.
70 c1zn1L_
not modelled
33.9
21
PDB header: biosynthetic/structural protein/rnaChain: L: PDB Molecule: 30s ribosomal protein s12;PDBTitle: coordinates of rrf fitted into cryo-em map of the 70s post-termination2 complex
71 c4abxB_
not modelled
33.1
12
PDB header: dna binding proteinChain: B: PDB Molecule: dna repair protein recn;PDBTitle: crystal structure of deinococcus radiodurans recn coiled-2 coil domain
72 c1wncE_
not modelled
32.6
20
PDB header: viral proteinChain: E: PDB Molecule: e2 glycoprotein;PDBTitle: crystal structure of the sars-cov spike protein fusion core
73 c5yfpG_
not modelled
32.2
14
PDB header: exocytosisChain: G: PDB Molecule: exocyst complex component exo70;PDBTitle: cryo-em structure of the exocyst complex
74 c1quuA_
not modelled
31.5
14
PDB header: contractile proteinChain: A: PDB Molecule: human skeletal muscle alpha-actinin 2;PDBTitle: crystal structure of two central spectrin-like repeats from alpha-2 actinin
75 c5j2lB_
not modelled
31.3
14
PDB header: de novo proteinChain: B: PDB Molecule: protein design 2l4hc2_11;PDBTitle: de novo design of protein homo-oligomers with modular hydrogen bond2 network-mediated specificity
76 c5c3lC_
not modelled
30.8
14
PDB header: transport proteinChain: C: PDB Molecule: nucleoporin nup62;PDBTitle: structure of the metazoan nup62.nup58.nup54 nucleoporin complex.
77 c5x5bB_
not modelled
30.5
14
PDB header: viral proteinChain: B: PDB Molecule: spike glycoprotein;PDBTitle: prefusion structure of sars-cov spike glycoprotein, conformation 2
78 c6nzkB_
not modelled
29.3
10
PDB header: viral proteinChain: B: PDB Molecule: spike surface glycoprotein;PDBTitle: structural basis for human coronavirus attachment to sialic acid2 receptors
79 c6ewyA_
not modelled
29.2
11
PDB header: structural proteinChain: A: PDB Molecule: peptidoglycan endopeptidase ripa;PDBTitle: ripa peptidoglycan hydrolase (rv1477, mycobacterium tuberculosis) n-2 terminal domain
80 c5yfpB_
not modelled
29.0
14
PDB header: exocytosisChain: B: PDB Molecule: exocyst complex component sec5;PDBTitle: cryo-em structure of the exocyst complex
81 c6cs2A_
not modelled
28.4
13
PDB header: viral protein/hydrolaseChain: A: PDB Molecule: spike glycoprotein,fibritin;PDBTitle: sars spike glycoprotein - human ace2 complex, stabilized variant, all2 ace2-bound particles
82 d2qall1
not modelled
27.7
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
83 c5ew5C_
not modelled
27.6
8
PDB header: hydrolaseChain: C: PDB Molecule: colicin-e9;PDBTitle: crystal structure of colicin e9 in complex with its immunity protein2 im9
84 c2bezC_
not modelled
27.2
11
PDB header: viral proteinChain: C: PDB Molecule: e2 glycoprotein;PDBTitle: structure of a proteolitically resistant core from the severe acute2 respiratory syndrome coronavirus s2 fusion protein
85 d1i94l_
not modelled
27.1
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
86 c2gl2B_
not modelled
27.1
11
PDB header: cell adhesionChain: B: PDB Molecule: adhesion a;PDBTitle: crystal structure of the tetra muntant (t66g,r67g,f68g,y69g) of2 bacterial adhesin fada
87 d1hcia4
not modelled
26.8
9
Fold: Spectrin repeat-likeSuperfamily: Spectrin repeatFamily: Spectrin repeat
88 c5i08A_
not modelled
26.5
14
PDB header: viral proteinChain: A: PDB Molecule: spike glycoprotein, envelope glycoprotein chimera;PDBTitle: prefusion structure of a human coronavirus spike protein
89 d2cp6a1
not modelled
26.2
23
Fold: SH3-like barrelSuperfamily: Cap-Gly domainFamily: Cap-Gly domain
90 c5xlrC_
not modelled
25.9
14
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: structure of sars-cov spike glycoprotein
91 c4wsrA_
not modelled
24.7
18
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin;PDBTitle: the crystal structure of hemagglutinin form a/chicken/new york/14677-2 13/1998
92 c5nugB_
not modelled
24.7
13
PDB header: motor proteinChain: B: PDB Molecule: cytoplasmic dynein 1 heavy chain 1;PDBTitle: motor domains from human cytoplasmic dynein-1 in the phi-particle2 conformation
93 d1fr3a_
not modelled
24.6
18
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP
94 c3ghgK_
not modelled
23.8
8
PDB header: blood clottingChain: K: PDB Molecule: fibrinogen beta chain;PDBTitle: crystal structure of human fibrinogen
95 c2q13A_
not modelled
23.6
9
PDB header: protein transportChain: A: PDB Molecule: dcc-interacting protein 13 alpha;PDBTitle: crystal structure of bar-ph domain of appl1
96 c1cz5A_
not modelled
21.9
12
PDB header: hydrolaseChain: A: PDB Molecule: vcp-like atpase;PDBTitle: nmr structure of vat-n: the n-terminal domain of vat (vcp-2 like atpase of thermoplasma)
97 c1yvlB_
not modelled
21.8
9
PDB header: signaling proteinChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: structure of unphosphorylated stat1
98 c5gasN_
not modelled
21.8
14
PDB header: hydrolaseChain: N: PDB Molecule: archaeal/vacuolar-type h+-atpase subunit i;PDBTitle: thermus thermophilus v/a-atpase, conformation 2
99 c1bg1A_
not modelled
21.6
10
PDB header: transcription/dnaChain: A: PDB Molecule: protein (transcription factor stat3b);PDBTitle: transcription factor stat3b/dna complex
100 c4iogD_
not modelled
21.3
11
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
101 c5szsC_
not modelled
21.2
12
PDB header: viral proteinChain: C: PDB Molecule: spike glycoprotein;PDBTitle: glycan shield and epitope masking of a coronavirus spike protein2 observed by cryo-electron microscopy
102 c3qr8A_
not modelled
20.7
15
PDB header: viral proteinChain: A: PDB Molecule: baseplate assembly protein v;PDBTitle: crystal structure of the bacteriophage p2 membrane-piercing protein2 gpv
103 c5cwsC_
not modelled
20.6
17
PDB header: protein transportChain: C: PDB Molecule: nucleoporin nsp1;PDBTitle: crystal structure of the intact chaetomium thermophilum nsp1-nup49-2 nup57 channel nucleoporin heterotrimer bound to its nic96 nuclear3 pore complex attachment site
104 c5n9yB_
not modelled
20.3
13
PDB header: membrane proteinChain: B: PDB Molecule: zinc transport protein zntb;PDBTitle: the full-length structure of zntb