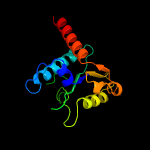
| 1 | c5mq9A_
|
|
 |
100.0 |
24 |
PDB header:translation
Chain: A: PDB Molecule:uncharacterized protein yacp;
PDBTitle: crystal structure of rae1 (yacp) from bacillus subtilis (w164l mutant)
|
|
|
|
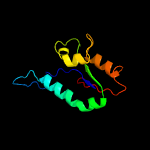
| 2 | d1cmwa2
|
|
 |
97.6 |
25 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
|
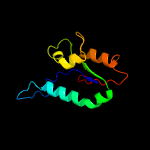
| 3 | c1cmwA_
|
|
 |
97.4 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:protein (dna polymerase i);
PDBTitle: crystal structure of taq dna-polymerase shows a new orientation for2 the structure-specific nuclease domain
|
|
|
|
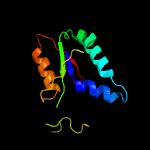
| 4 | c3zddA_
|
|
 |
97.4 |
23 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:protein xni;
PDBTitle: structure of e. coli exoix in complex with the palindromic 5ov62 oligonucleotide and potassium
|
|
|
|
| 5 | c6c34A_
|
|
 |
97.3 |
18 |
PDB header:dna binding protein
Chain: A: PDB Molecule:5'-3' exonuclease;
PDBTitle: mycobacterium smegmatis dna flap endonuclease mutant d125n
|
|
|
|
| 6 | c1ut8B_
|
|
 |
97.2 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:exodeoxyribonuclease;
PDBTitle: divalent metal ions (zinc) bound to t5 5'-exonuclease
|
|
|
|
| 7 | d1tfra2
|
|
 |
97.0 |
17 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
|
| 8 | d1xo1a2
|
|
 |
96.9 |
21 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
|
| 9 | c3v32B_
|
|
 |
95.9 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:ribonuclease zc3h12a;
PDBTitle: crystal structure of mcpip1 n-terminal conserved domain
|
|
|
|
| 10 | c3v33A_
|
|
 |
95.6 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribonuclease zc3h12a;
PDBTitle: crystal structure of mcpip1 conserved domain with zinc-finger motif
|
|
|
|
| 11 | c3ix7A_
|
|
 |
94.6 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ttha0540;
PDBTitle: crystal structure of a domain of functionally unknown protein from2 thermus thermophilus hb8
|
|
|
|
| 12 | c2izoA_
|
|
 |
94.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap structure-specific endonuclease;
PDBTitle: structure of an archaeal pcna1-pcna2-fen1 complex
|
|
|
|
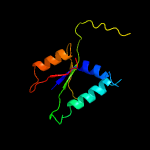
| 13 | d1ul1x2
|
|
 |
93.7 |
18 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
|
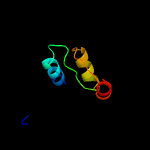
| 14 | c2hwwC_
|
|
 |
93.1 |
20 |
PDB header:rna binding protein
Chain: C: PDB Molecule:telomerase-binding protein est1a;
PDBTitle: structure of pin domain of human smg6
|
|
|
|
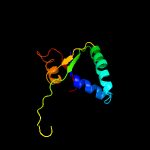
| 15 | c1b43A_
|
|
 |
92.2 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:protein (fen-1);
PDBTitle: fen-1 from p. furiosus
|
|
|
|
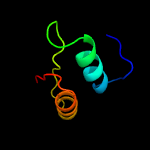
| 16 | d1o4wa_
|
|
 |
91.9 |
17 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:PIN domain |
|
|
|
| 17 | c5jpqd_
|
|
 |
91.7 |
33 |
PDB header:ribosome
Chain: D: PDB Molecule:wd40 domain proteins;
PDBTitle: cryo-em structure of the 90s pre-ribosome
|
|
|
|
| 18 | c1a77A_
|
|
 |
90.5 |
25 |
PDB header:5'-3' exo/endo nuclease
Chain: A: PDB Molecule:flap endonuclease-1 protein;
PDBTitle: flap endonuclease-1 from methanococcus jannaschii
|
|
|
|
| 19 | c2ihnA_
|
|
 |
90.3 |
18 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:ribonuclease h;
PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
|
|
|
|
| 20 | c4wa8A_
|
|
 |
89.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: methanopyrus kandleri fen-1 nuclease
|
|
|
|
| 21 | c5yz4A_ |
|
not modelled |
89.2 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:rrna-processing protein fcf1;
PDBTitle: structure of the pin domain endonuclease utp24
|
|
|
| 22 | c3q8lA_ |
|
not modelled |
89.0 |
21 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: crystal structure of human flap endonuclease fen1 (wt) in complex with2 substrate 5'-flap dna, sm3+, and k+
|
|
|
| 23 | c1ul1Y_ |
|
not modelled |
88.0 |
18 |
PDB header:hydrolase/dna binding protein
Chain: Y: PDB Molecule:flap endonuclease-1;
PDBTitle: crystal structure of the human fen1-pcna complex
|
|
|
| 24 | c4mj7B_ |
|
not modelled |
88.0 |
22 |
PDB header:rna binding protein
Chain: B: PDB Molecule:rrna-processing protein utp23;
PDBTitle: crystal structure of the pin domain of saccharomyces cerevisiae utp23
|
|
|
| 25 | c1rxvA_ |
|
not modelled |
87.0 |
28 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:flap structure-specific endonuclease;
PDBTitle: crystal structure of a. fulgidus fen-1 bound to dna
|
|
|
| 26 | c3i8oA_ |
|
not modelled |
79.9 |
18 |
PDB header:rna binding protein
Chain: A: PDB Molecule:kh domain-containing protein mj1533;
PDBTitle: a domain of a functionally unknown protein from methanocaldococcus2 jannaschii dsm 2661.
|
|
|
| 27 | c4q0rB_ |
|
not modelled |
65.2 |
19 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:dna repair protein rad2;
PDBTitle: the catalytic core of rad2 (complex i)
|
|
|
| 28 | c3qeaZ_ |
|
not modelled |
61.3 |
14 |
PDB header:hydrolase/dna
Chain: Z: PDB Molecule:exonuclease 1;
PDBTitle: crystal structure of human exonuclease 1 exo1 (wt) in complex with dna2 (complex ii)
|
|
|
| 29 | d1a77a2 |
|
not modelled |
58.6 |
26 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 30 | c5f4hF_ |
|
not modelled |
55.2 |
13 |
PDB header:hydrolase
Chain: F: PDB Molecule:nucleotide binding protein pinc;
PDBTitle: archael ruvb-like holiday junction helicase
|
|
|
| 31 | d1y82a1 |
|
not modelled |
50.1 |
14 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:PIN domain |
|
|
| 32 | c2zktB_ |
|
not modelled |
48.4 |
17 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: structure of ph0037 protein from pyrococcus horikoshii
|
|
|
| 33 | c5ywwA_ |
|
not modelled |
45.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleotide binding protein pinc;
PDBTitle: archael ruvb-like holiday junction helicase
|
|
|
| 34 | d1o98a2 |
|
not modelled |
44.3 |
20 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain |
|
|
| 35 | d1rxwa2 |
|
not modelled |
40.6 |
21 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 36 | d1pkla3 |
|
not modelled |
27.1 |
24 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 37 | c5t9jB_ |
|
not modelled |
26.7 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:flap endonuclease gen homolog 1;
PDBTitle: crystal structure of human gen1 in complex with holliday junction dna2 in the upper interface
|
|
|
| 38 | c2ayxA_ |
|
not modelled |
24.4 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:sensor kinase protein rcsc;
PDBTitle: solution structure of the e.coli rcsc c-terminus (residues2 700-949) containing linker region and phosphoreceiver3 domain
|
|
|
| 39 | c3m8yC_ |
|
not modelled |
24.2 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:phosphopentomutase;
PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
|
|
|
| 40 | c2i09A_ |
|
not modelled |
23.6 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphopentomutase;
PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
|
|
|
| 41 | c3qoyA_ |
|
not modelled |
22.4 |
15 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:50s ribosomal protein l1;
PDBTitle: crystal structure of ribosomal protein l1 from aquifex aeolicus
|
|
|
| 42 | c3hdvB_ |
|
not modelled |
20.2 |
18 |
PDB header:transcription regulator
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of response regulator receiver protein from2 pseudomonas putida
|
|
|
| 43 | d1liua3 |
|
not modelled |
20.1 |
15 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 44 | d1e0ta3 |
|
not modelled |
19.6 |
21 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 45 | c3lxqB_ |
|
not modelled |
19.3 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein vp1736;
PDBTitle: the crystal structure of a protein in the alkaline phosphatase2 superfamily from vibrio parahaemolyticus to 1.95a
|
|
|
| 46 | c4o1iA_ |
|
not modelled |
19.2 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: crystal structure of the regulatory domain of mtbglnr
|
|
|
| 47 | d2g50a3 |
|
not modelled |
19.1 |
18 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 48 | c4my4A_ |
|
not modelled |
19.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: crystal structure of phosphoglycerate mutase from staphylococcus2 aureus.
|
|
|
| 49 | d1a3xa3 |
|
not modelled |
17.7 |
18 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 50 | d1yioa2 |
|
not modelled |
17.4 |
20 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 51 | c3tb4A_ |
|
not modelled |
16.9 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:vibriobactin-specific isochorismatase;
PDBTitle: crystal structure of the isc domain of vibb
|
|
|
| 52 | c6m8oA_ |
|
not modelled |
16.0 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:dna-binding response regulator;
PDBTitle: crystal structure of the receiver domain of lytr from staphylococcus2 aureus
|
|
|
| 53 | c1o98A_ |
|
not modelled |
16.0 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent
PDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
|
|
|
| 54 | d1p49a_ |
|
not modelled |
15.7 |
17 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 55 | c3bboD_ |
|
not modelled |
15.6 |
20 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein l1;
PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
|
|
|
| 56 | c5t3yA_ |
|
not modelled |
15.5 |
25 |
PDB header:signaling protein
Chain: A: PDB Molecule:two-component system response regulator;
PDBTitle: solution structure of response regulator protein from burkholderia2 multivorans
|
|
|
| 57 | c3lteH_ |
|
not modelled |
15.5 |
16 |
PDB header:transcription
Chain: H: PDB Molecule:response regulator;
PDBTitle: crystal structure of response regulator (signal receiver domain) from2 bermanella marisrubri
|
|
|
| 58 | d1pcva_ |
|
not modelled |
15.4 |
57 |
Fold:Osmotin, thaumatin-like protein
Superfamily:Osmotin, thaumatin-like protein
Family:Osmotin, thaumatin-like protein |
|
|
| 59 | d1v96a1 |
|
not modelled |
15.3 |
17 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:PIN domain |
|
|
| 60 | c3oryA_ |
|
not modelled |
15.1 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: crystal structure of flap endonuclease 1 from hyperthermophilic2 archaeon desulfurococcus amylolyticus
|
|
|
| 61 | c3igzB_ |
|
not modelled |
15.0 |
21 |
PDB header:isomerase
Chain: B: PDB Molecule:cofactor-independent phosphoglycerate mutase;
PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
|
|
|
| 62 | c2gyc2_ |
|
not modelled |
15.0 |
22 |
PDB header:ribosome
Chain: 2: PDB Molecule:50s ribosomal protein l1;
PDBTitle: structure of the 50s subunit of a secm-stalled e. coli ribosome2 complex obtained by fitting atomic models for rna and protein3 components into cryo-em map emd-1143
|
|
|
| 63 | d1d8wa_ |
|
not modelled |
14.7 |
37 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:L-rhamnose isomerase |
|
|
| 64 | d1du5a_ |
|
not modelled |
14.4 |
57 |
Fold:Osmotin, thaumatin-like protein
Superfamily:Osmotin, thaumatin-like protein
Family:Osmotin, thaumatin-like protein |
|
|
| 65 | d1auka_ |
|
not modelled |
14.0 |
16 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 66 | c2rdmB_ |
|
not modelled |
13.9 |
13 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver protein from2 sinorhizobium medicae wsm419
|
|
|
| 67 | c2ahnA_ |
|
not modelled |
13.7 |
57 |
PDB header:allergen
Chain: A: PDB Molecule:thaumatin-like protein;
PDBTitle: high resolution structure of a cherry allergen pru av 2
|
|
|
| 68 | c3hheA_ |
|
not modelled |
13.5 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from bartonella2 henselae
|
|
|
| 69 | d1fsua_ |
|
not modelled |
12.8 |
8 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 70 | d1hdha_ |
|
not modelled |
12.7 |
25 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 71 | c5cnqA_ |
|
not modelled |
12.3 |
17 |
PDB header:replication
Chain: A: PDB Molecule:nuclease-like protein;
PDBTitle: crystal structure of the holliday junction-resolving enzyme gen1 (wt)2 in complex with product dna, mg2+ and mn2+ ions
|
|
|
| 72 | c5vpuA_ |
|
not modelled |
12.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: crystal structure of 2,3-bisphosphoglycerate-independent2 phosphoglycerate mutase bound to 3-phosphoglycerate, from3 acinetobacter baumannii
|
|
|
| 73 | d1auna_ |
|
not modelled |
11.8 |
43 |
Fold:Osmotin, thaumatin-like protein
Superfamily:Osmotin, thaumatin-like protein
Family:Osmotin, thaumatin-like protein |
|
|
| 74 | c3qtgA_ |
|
not modelled |
11.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:pyruvate kinase;
PDBTitle: crystal structure of pyruvate kinase from pyrobaculum aerophilum
|
|
|
| 75 | c3p14C_ |
|
not modelled |
11.6 |
41 |
PDB header:isomerase
Chain: C: PDB Molecule:l-rhamnose isomerase;
PDBTitle: crystal structure of l-rhamnose isomerase with a novel high thermo-2 stability from bacillus halodurans
|
|
|
| 76 | c5k4pA_ |
|
not modelled |
11.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:probable phosphatidylethanolamine transferase mcr-1;
PDBTitle: catalytic domain of mcr-1 phosphoethanolamine transferase
|
|
|
| 77 | c5kgmA_ |
|
not modelled |
11.6 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: 2.95a resolution structure of apo independent phosphoglycerate mutase2 from c. elegans (monoclinic form)
|
|
|
| 78 | c5fgnA_ |
|
not modelled |
11.5 |
20 |
PDB header:transferase,hydrolase
Chain: A: PDB Molecule:lipooligosaccharide phosphoethanolamine transferase a;
PDBTitle: integral membrane protein lipooligosaccharide phosphoethanolamine2 transferase a (epta) from neisseria meningitidis
|
|
|
| 79 | c6a82A_ |
|
not modelled |
11.5 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoethanolamine transferase eptc;
PDBTitle: crystal structure of the c-terminal periplasmic domain of eceptc from2 escherichia coli
|
|
|
| 80 | c5i5fA_ |
|
not modelled |
11.3 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein yejm;
PDBTitle: salmonella global domain 191
|
|
|
| 81 | d1vh3a_ |
|
not modelled |
11.3 |
14 |
Fold:Nucleotide-diphospho-sugar transferases
Superfamily:Nucleotide-diphospho-sugar transferases
Family:Cytidylytransferase |
|
|
| 82 | c3ed4A_ |
|
not modelled |
11.2 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:arylsulfatase;
PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
|
|
|
| 83 | d2i09a1 |
|
not modelled |
11.0 |
13 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:DeoB catalytic domain-like |
|
|
| 84 | d2a9pa1 |
|
not modelled |
10.9 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 85 | d1l7ba_ |
|
not modelled |
10.8 |
14 |
Fold:BRCT domain
Superfamily:BRCT domain
Family:DNA ligase |
|
|
| 86 | c2w8dB_ |
|
not modelled |
10.4 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:processed glycerol phosphate lipoteichoic acid synthase 2;
PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
|
|
|
| 87 | c3b5qB_ |
|
not modelled |
9.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of a putative sulfatase (np_810509.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.40 a resolution
|
|
|
| 88 | c1ew2A_ |
|
not modelled |
9.9 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphatase;
PDBTitle: crystal structure of a human phosphatase
|
|
|
| 89 | d1zeda1 |
|
not modelled |
9.9 |
28 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Alkaline phosphatase |
|
|
| 90 | c4qpjC_ |
|
not modelled |
9.8 |
11 |
PDB header:signaling protein/dna binding protein
Chain: C: PDB Molecule:cell cycle response regulator ctra;
PDBTitle: 2.7 angstrom structure of a phosphotransferase in complex with a2 receiver domain
|
|
|
| 91 | c1t5aB_ |
|
not modelled |
9.8 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase, m2 isozyme;
PDBTitle: human pyruvate kinase m2
|
|
|
| 92 | c5jyuA_ |
|
not modelled |
9.5 |
14 |
PDB header:sigaling protein
Chain: A: PDB Molecule:two-component sensor histidine kinase;
PDBTitle: nmr structure of pseudo receiver domain of cika from2 thermosynechococcus elongatus
|
|
|
| 93 | c3q3qA_ |
|
not modelled |
9.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase;
PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
|
|
|
| 94 | c3ktoA_ |
|
not modelled |
9.1 |
12 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver protein2 from pseudoalteromonas atlantica
|
|
|
| 95 | c3a52A_ |
|
not modelled |
9.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:cold-active alkaline phosphatase;
PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
|
|
|
| 96 | d1ad2a_ |
|
not modelled |
9.1 |
12 |
Fold:Ribosomal protein L1
Superfamily:Ribosomal protein L1
Family:Ribosomal protein L1 |
|
|
| 97 | c2l69A_ |
|
not modelled |
9.0 |
25 |
PDB header:de novo protein
Chain: A: PDB Molecule:rossmann 2x3 fold protein;
PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or28
|
|
|
| 98 | c4uopB_ |
|
not modelled |
9.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:lipoteichoic acid primase;
PDBTitle: crystal structure of the lipoteichoic acid synthase ltap from listeria2 monocytogenes
|
|
|
| 99 | c4fdiA_ |
|
not modelled |
8.9 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylgalactosamine-6-sulfatase;
PDBTitle: the molecular basis of mucopolysaccharidosis iv a
|
|
|
















































































































