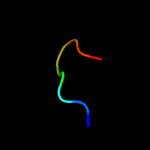| 1 |
|
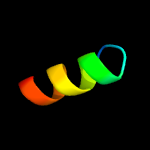
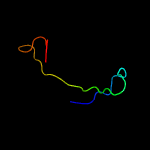
PDB 5e9f chain D
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 18.1%
Identity: 44%
PDB header:lyase
Chain: D: PDB Molecule:isocitrate lyase;
PDBTitle: structural insights of isocitrate lyases from magnaporthe oryzae
Phyre2
| 2 |
|
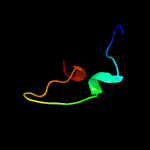
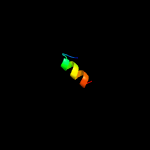
PDB 2r7m chain A
Region: 93 - 114
Aligned: 22
Modelled: 22
Confidence: 14.9%
Identity: 50%
PDB header:ligase
Chain: A: PDB Molecule:5-formaminoimidazole-4-carboxamide-1-(beta)-d-ribofuranosyl
PDBTitle: crystal structure of faicar synthetase (purp) from m. jannaschii2 complexed with amp
Phyre2
| 3 |
|
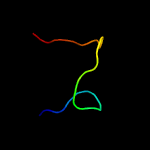
PDB 5d5o chain E
Region: 32 - 40
Aligned: 9
Modelled: 9
Confidence: 13.4%
Identity: 67%
PDB header:unknown function
Chain: E: PDB Molecule:uncharacterized protein mj0489;
PDBTitle: hcgc from methanocaldococcus jannaschii
Phyre2
| 4 |
|
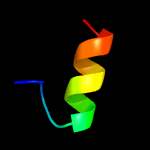
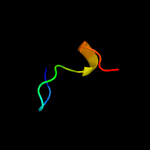
PDB 1dqu chain A
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 13.0%
Identity: 44%
Fold: TIM beta/alpha-barrel
Superfamily: Phosphoenolpyruvate/pyruvate domain
Family: Phosphoenolpyruvate mutase/Isocitrate lyase-like
Phyre2
| 5 |
|
PDB 3bne chain A domain 2
Region: 10 - 43
Aligned: 33
Modelled: 34
Confidence: 12.2%
Identity: 21%
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)
Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)
Family: Lipoxigenase N-terminal domain
Phyre2
| 6 |
|
PDB 1f61 chain A
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 11.9%
Identity: 44%
Fold: TIM beta/alpha-barrel
Superfamily: Phosphoenolpyruvate/pyruvate domain
Family: Phosphoenolpyruvate mutase/Isocitrate lyase-like
Phyre2
| 7 |
|
PDB 1igw chain A
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 11.6%
Identity: 31%
Fold: TIM beta/alpha-barrel
Superfamily: Phosphoenolpyruvate/pyruvate domain
Family: Phosphoenolpyruvate mutase/Isocitrate lyase-like
Phyre2
| 8 |
|
PDB 3j81 chain J
Region: 64 - 84
Aligned: 21
Modelled: 21
Confidence: 11.5%
Identity: 24%
PDB header:ribosome
Chain: J: PDB Molecule:us4;
PDBTitle: cryoem structure of a partial yeast 48s preinitiation complex
Phyre2
| 9 |
|
PDB 5o4j chain C
Region: 32 - 40
Aligned: 9
Modelled: 9
Confidence: 10.5%
Identity: 67%
PDB header:transferase
Chain: C: PDB Molecule:hcgc;
PDBTitle: hcgc from methanococcus maripaludis cocrystallized with sah and2 pyridinol
Phyre2
| 10 |
|
PDB 2j01 chain D domain 1
Region: 101 - 116
Aligned: 15
Modelled: 16
Confidence: 9.3%
Identity: 47%
Fold: SH3-like barrel
Superfamily: Translation proteins SH3-like domain
Family: C-terminal domain of ribosomal protein L2
Phyre2
| 11 |
|
PDB 2qam chain C domain 1
Region: 101 - 116
Aligned: 15
Modelled: 16
Confidence: 8.8%
Identity: 33%
Fold: SH3-like barrel
Superfamily: Translation proteins SH3-like domain
Family: C-terminal domain of ribosomal protein L2
Phyre2
| 12 |
|
PDB 2gya chain A
Region: 101 - 108
Aligned: 8
Modelled: 8
Confidence: 7.9%
Identity: 38%
PDB header:ribosome
Chain: A: PDB Molecule:50s ribosomal protein l2;
PDBTitle: structure of the 50s subunit of a pre-translocational e. coli ribosome2 obtained by fitting atomic models for rna and protein components into3 cryo-em map emd-1056
Phyre2
| 13 |
|
PDB 2zjr chain A domain 1
Region: 101 - 108
Aligned: 8
Modelled: 8
Confidence: 7.8%
Identity: 50%
Fold: SH3-like barrel
Superfamily: Translation proteins SH3-like domain
Family: C-terminal domain of ribosomal protein L2
Phyre2
| 14 |
|
PDB 1q46 chain A
Region: 64 - 85
Aligned: 22
Modelled: 22
Confidence: 7.2%
Identity: 23%
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor 2 alpha subunit;
PDBTitle: crystal structure of the eif2 alpha subunit from2 saccharomyces cerevisia
Phyre2
| 15 |
|
PDB 3e5b chain B
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 6.8%
Identity: 25%
PDB header:lyase
Chain: B: PDB Molecule:isocitrate lyase;
PDBTitle: 2.4 a crystal structure of isocitrate lyase from brucella melitensis
Phyre2
| 16 |
|
PDB 3i4e chain A
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 6.4%
Identity: 31%
PDB header:lyase
Chain: A: PDB Molecule:isocitrate lyase;
PDBTitle: crystal structure of isocitrate lyase from burkholderia2 pseudomallei
Phyre2
| 17 |
|
PDB 1inz chain A
Region: 70 - 108
Aligned: 39
Modelled: 39
Confidence: 6.3%
Identity: 33%
Fold: alpha-alpha superhelix
Superfamily: ENTH/VHS domain
Family: ENTH domain
Phyre2
| 18 |
|
PDB 3d5b chain D
Region: 101 - 116
Aligned: 15
Modelled: 16
Confidence: 6.3%
Identity: 47%
PDB header:ribosome
Chain: D: PDB Molecule:50s ribosomal protein l2;
PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of one 70s ribosome. the entire crystal3 structure contains two 70s ribosomes as described in remark 400.
Phyre2
| 19 |
|
PDB 2z4l chain C
Region: 101 - 108
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 38%
PDB header:ribosome
Chain: C: PDB Molecule:50s ribosomal protein l2;
PDBTitle: crystal structure of the bacterial ribosome from escherichia coli in2 complex with paromomycin and ribosome recycling factor (rrf). this3 file contains the 50s subunit of the first 70s ribosome, with4 paromomycin and rrf bound. the entire crystal structure contains two5 70s ribosomes and is described in remark 400.
Phyre2
| 20 |
|
PDB 4owf chain G
Region: 9 - 14
Aligned: 6
Modelled: 6
Confidence: 5.8%
Identity: 83%
PDB header:transcription/protein binding
Chain: G: PDB Molecule:e3 ubiquitin-protein ligase rnf31;
PDBTitle: crystal structure of the nemo cozi in complex with hoip nzf1 domain
Phyre2
| 21 |
|