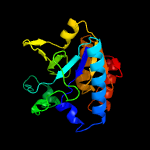
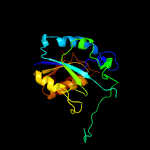
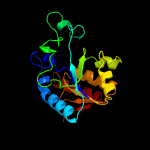
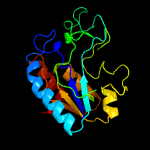
1 c3gbcA_
100.0
100
PDB header: hydrolaseChain: A: PDB Molecule: pyrazinamidase/nicotinamidas pnca;PDBTitle: determination of the crystal structure of the pyrazinamidase from2 m.tuberculosis : a structure-function analysis for prediction3 resistance to pyrazinamide
2 d1nbaa_
100.0
20
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
3 c2fq1A_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: isochorismatase;PDBTitle: crystal structure of the two-domain non-ribosomal peptide synthetase2 entb containing isochorismate lyase and aryl-carrier protein domains
4 c2h0rD_
100.0
29
PDB header: hydrolaseChain: D: PDB Molecule: nicotinamidase;PDBTitle: structure of the yeast nicotinamidase pnc1p
5 c3hu5B_
100.0
27
PDB header: hydrolaseChain: B: PDB Molecule: isochorismatase family protein;PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
6 d1im5a_
100.0
36
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
7 c3irvA_
100.0
26
PDB header: hydrolaseChain: A: PDB Molecule: cysteine hydrolase;PDBTitle: crystal structure of cysteine hydrolase pspph_2384 from pseudomonas2 syringae pv. phaseolicola 1448a
8 c3ot4F_
100.0
26
PDB header: hydrolaseChain: F: PDB Molecule: putative isochorismatase;PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
9 c5gleA_
100.0
18
PDB header: lyaseChain: A: PDB Molecule: ischorismate lyase;PDBTitle: the structure of vibrio anguillarum775 angb-icl
10 c3r2jC_
100.0
40
PDB header: hydrolaseChain: C: PDB Molecule: alpha/beta-hydrolase-like protein;PDBTitle: crystal structure of pnc1 from l. infantum in complex with nicotinate
11 d1nf9a_
100.0
18
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
12 c3hb7G_
100.0
25
PDB header: hydrolaseChain: G: PDB Molecule: isochorismatase hydrolase;PDBTitle: the crystal structure of an isochorismatase-like hydrolase from2 alkaliphilus metalliredigens to 2.3a
13 c5zn8B_
100.0
26
PDB header: metal binding proteinChain: B: PDB Molecule: isochorismatase;PDBTitle: crystal structure of nicotinamidase pnca from bacillus subtilis
14 c5wxvL_
100.0
19
PDB header: lyaseChain: L: PDB Molecule: isochorismate lyase;PDBTitle: the crystal structure of vabb-icl domain from vibrio anguillarum 775
15 d1j2ra_
100.0
18
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
16 c6azoC_
100.0
24
PDB header: hydrolaseChain: C: PDB Molecule: putative amidase;PDBTitle: structural and biochemical characterization of a non-canonical biuret2 hydrolase (biuh) from the cyanuric acid catabolism pathway of3 rhizobium leguminasorum bv. viciae 3841
17 c3tb4A_
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: vibriobactin-specific isochorismatase;PDBTitle: crystal structure of the isc domain of vibb
18 c3o93A_
100.0
26
PDB header: hydrolaseChain: A: PDB Molecule: nicotinamidase;PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
19 c3eefA_
100.0
28
PDB header: hydrolaseChain: A: PDB Molecule: n-carbamoylsarcosine amidase related protein;PDBTitle: crystal structure of n-carbamoylsarcosine amidase from thermoplasma2 acidophilum
20 c2wtaA_
100.0
37
PDB header: hydrolaseChain: A: PDB Molecule: nicotinamidase;PDBTitle: acinetobacter baumanii nicotinamidase pyrazinamidease
21 c3oqpA_
not modelled
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: putative isochorismatase;PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
22 c3oqpB_
not modelled
100.0
24
PDB header: hydrolaseChain: B: PDB Molecule: putative isochorismatase;PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
23 c3kl2K_
not modelled
100.0
21
PDB header: structural genomics, unknown functionChain: K: PDB Molecule: putative isochorismatase;PDBTitle: crystal structure of a putative isochorismatase from streptomyces2 avermitilis
24 c3mcwA_
not modelled
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of an a putative hydrolase of the isochorismatase2 family (cv_1320) from chromobacterium violaceum atcc 12472 at 1.06 a3 resolution
25 c4h17B_
not modelled
100.0
19
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase, isochorismatase family;PDBTitle: crystal structure of an isochorismatase (pp1826) from pseudomonas2 putida kt2440 at 1.60 a resolution
26 c2a67C_
not modelled
100.0
19
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: isochorismatase family protein;PDBTitle: crystal structure of isochorismatase family protein
27 c3lqyA_
not modelled
100.0
25
PDB header: hydrolaseChain: A: PDB Molecule: putative isochorismatase hydrolase;PDBTitle: crystal structure of putative isochorismatase hydrolase from oleispira2 antarctica
28 c5ha8A_
not modelled
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: isochorismatase;PDBTitle: structure of a cysteine hydrolase
29 c1yzvA_
not modelled
100.0
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: hypothetical protein from trypanosoma cruzi
30 d1yaca_
not modelled
100.0
17
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
31 c2b34C_
not modelled
100.0
23
PDB header: hydrolaseChain: C: PDB Molecule: mar1 ribonuclease;PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
32 d1x9ga_
not modelled
100.0
18
Fold: Isochorismatase-like hydrolasesSuperfamily: Isochorismatase-like hydrolasesFamily: Isochorismatase-like hydrolases
33 c5kinD_
not modelled
87.0
11
PDB header: lyaseChain: D: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
34 c1ddzA_
not modelled
85.2
12
PDB header: lyaseChain: A: PDB Molecule: carbonic anhydrase;PDBTitle: x-ray structure of a beta-carbonic anhydrase from the red2 alga, porphyridium purpureum r-1
35 d1vp8a_
not modelled
82.1
26
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like
36 d1xdpa3
not modelled
74.5
24
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain
37 d1pv8a_
not modelled
74.4
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
38 c1x1qA_
not modelled
73.2
17
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase beta chain from thermus2 thermophilus hb8
39 c4negA_
not modelled
64.3
13
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: the crystal structure of tryptophan synthase subunit beta from2 bacillus anthracis str. 'ames ancestor'
40 d1cmwa2
not modelled
59.6
16
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain
41 c1xdoB_
not modelled
59.5
22
PDB header: transferaseChain: B: PDB Molecule: polyphosphate kinase;PDBTitle: crystal structure of escherichia coli polyphosphate kinase
42 d1onfa2
not modelled
58.9
10
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
43 d1oi7a1
not modelled
57.6
21
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
44 d1f2da_
not modelled
54.5
10
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
45 d1tfra2
not modelled
54.2
7
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: 5' to 3' exonuclease catalytic domain
46 d1ae1a_
not modelled
54.0
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
47 d2ae2a_
not modelled
52.2
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
48 c2ihnA_
not modelled
52.1
5
PDB header: hydrolase/dnaChain: A: PDB Molecule: ribonuclease h;PDBTitle: co-crystal of bacteriophage t4 rnase h with a fork dna2 substrate
49 d1qopb_
not modelled
51.0
18
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
50 c5yvmA_
not modelled
50.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: alcohol dehydrogenase;PDBTitle: crystal structure of the archaeal halo-thermophilic red sea brine pool2 alcohol dehydrogenase adh/d1 bound to nzq
51 c4qysA_
not modelled
49.1
11
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain 2;PDBTitle: trpb2 enzymes
52 d1v8za1
not modelled
48.3
11
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
53 d1qwga_
not modelled
45.0
12
Fold: TIM beta/alpha-barrelSuperfamily: (2r)-phospho-3-sulfolactate synthase ComAFamily: (2r)-phospho-3-sulfolactate synthase ComA
54 c4jcmA_
not modelled
44.1
10
PDB header: transferaseChain: A: PDB Molecule: cyclodextrin glucanotransferase;PDBTitle: crystal structure of gamma-cgtase from alkalophilic bacillus clarkii2 at 1.65 angstrom resolution
55 c2o2jA_
not modelled
43.1
23
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain;PDBTitle: mycobacterium tuberculosis tryptophan synthase beta chain dimer2 (apoform)
56 c5yw2D_
not modelled
42.8
23
PDB header: transferaseChain: D: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 francisella tularensis.
57 d1ddza1
not modelled
42.4
14
Fold: Resolvase-likeSuperfamily: beta-carbonic anhydrase, cabFamily: beta-carbonic anhydrase, cab
58 c6hpfA_
not modelled
40.7
19
PDB header: hydrolaseChain: A: PDB Molecule: endo-b-mannanase;PDBTitle: structure of inactive e165q mutant of fungal non-cbm carrying gh262 endo-b-mannanase from yunnania penicillata in complex with alpha-62-3 61-di-galactosyl-mannotriose
59 c5tchH_
not modelled
40.5
23
PDB header: lyaseChain: H: PDB Molecule: tryptophan synthase beta chain;PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
60 d1a9xa3
not modelled
39.5
17
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: BC N-terminal domain-like
61 c5yaaD_
not modelled
38.9
17
PDB header: hydrolaseChain: D: PDB Molecule: meiosis regulator and mrna stability factor 1;PDBTitle: crystal structure of marf1 nyn domain from mus musculus
62 c3zm8A_
not modelled
38.8
17
PDB header: hydrolaseChain: A: PDB Molecule: gh26 endo-beta-1,4-mannanase;PDBTitle: crystal structure of podospora anserina gh26-cbm352 beta-(1,4)-mannanase
63 c2wanA_
not modelled
38.7
11
PDB header: hydrolaseChain: A: PDB Molecule: pullulanase;PDBTitle: pullulanase from bacillus acidopullulyticus
64 c5ybwA_
not modelled
38.3
23
PDB header: isomeraseChain: A: PDB Molecule: aspartate racemase;PDBTitle: crystal structure of pyridoxal 5'-phosphate-dependent aspartate2 racemase
65 c6ci9D_
not modelled
38.1
15
PDB header: oxidoreductaseChain: D: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] reductase;PDBTitle: rmm microcompartment-associated aminopropanol dehydrogenase nadp +2 aminoacetone holo-structure
66 d1v7ca_
not modelled
38.0
13
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
67 c3toxG_
not modelled
37.9
19
PDB header: oxidoreductaseChain: G: PDB Molecule: short chain dehydrogenase;PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
68 c2zsjB_
not modelled
37.1
18
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
69 c5ig2B_
not modelled
36.7
19
PDB header: oxidoreductaseChain: B: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: crystal structure of a short chain dehydrogenase/reductase sdr from2 burkholderia phymatum in complex with nad
70 c3zddA_
not modelled
35.9
21
PDB header: hydrolase/dnaChain: A: PDB Molecule: protein xni;PDBTitle: structure of e. coli exoix in complex with the palindromic 5ov62 oligonucleotide and potassium
71 c6iaqA_
not modelled
35.4
10
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase n-terminus domain-containingPDBTitle: structure of amine dehydrogenase from mycobacterium smegmatis
72 c4yaiB_
not modelled
35.0
17
PDB header: oxidoreductaseChain: B: PDB Molecule: c alpha-dehydrogenase;PDBTitle: crystal structure of ligl in complex with nadh and gge from2 sphingobium sp. strain syk-6
73 c2dy0A_
not modelled
34.0
31
PDB header: transferaseChain: A: PDB Molecule: adenine phosphoribosyltransferase;PDBTitle: crystal structure of project jw0458 from escherichia coli
74 c2zatC_
not modelled
33.8
12
PDB header: oxidoreductaseChain: C: PDB Molecule: dehydrogenase/reductase sdr family member 4;PDBTitle: crystal structure of a mammalian reductase
75 c1cmwA_
not modelled
33.6
21
PDB header: transferaseChain: A: PDB Molecule: protein (dna polymerase i);PDBTitle: crystal structure of taq dna-polymerase shows a new orientation for2 the structure-specific nuclease domain
76 d1xp8a1
not modelled
33.2
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
77 d1j0aa_
not modelled
33.1
13
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
78 d2nu7a1
not modelled
33.0
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
79 d1nhpa2
not modelled
32.7
22
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
80 c6hulB_
not modelled
31.7
14
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase beta chain 1;PDBTitle: sulfolobus solfataricus tryptophan synthase ab complex
81 c4urfB_
not modelled
30.8
17
PDB header: oxidoreductaseChain: B: PDB Molecule: cyclohexanol dehydrogenase;PDBTitle: molecular genetic and crystal structural analysis of 1-(4-2 hydroxyphenyl)-ethanol dehydrogenase from aromatoleum aromaticum ebn1
82 c3euaD_
not modelled
30.3
15
PDB header: isomeraseChain: D: PDB Molecule: putative fructose-aminoacid-6-phosphate deglycase;PDBTitle: crystal structure of a putative phosphosugar isomerase (bsu32610) from2 bacillus subtilis at 1.90 a resolution
83 c3imfA_
not modelled
29.8
15
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain dehydrogenase;PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
84 d1o2da_
not modelled
29.7
16
Fold: Dehydroquinate synthase-likeSuperfamily: Dehydroquinate synthase-likeFamily: Iron-containing alcohol dehydrogenase
85 d1tyza_
not modelled
29.6
16
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
86 c6aioA_
not modelled
29.0
17
PDB header: flavoproteinChain: A: PDB Molecule: pnpa;PDBTitle: crystal structure of p-nitrophenol 4-monooxygenase pnpa from2 pseudomonas putida dll-e4
87 d1d7ya2
not modelled
29.0
15
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
88 c4j2hA_
not modelled
28.6
15
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain alcohol dehydrogenase-related dehydrogenase;PDBTitle: crystal structure of a putative short-chain alcohol dehydrogenase from2 sinorhizobium meliloti 1021 (target nysgrc-011708)
89 c5idxB_
not modelled
28.1
25
PDB header: oxidoreductaseChain: B: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: crystal structure of an oxidoreductase from burkholderia vietnamiensis
90 d1o58a_
not modelled
28.0
18
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
91 c5u9pB_
not modelled
27.7
9
PDB header: oxidoreductaseChain: B: PDB Molecule: gluconate 5-dehydrogenase;PDBTitle: crystal structure of a gluconate 5-dehydrogenase from burkholderia2 cenocepacia j2315 in complex with nadp and tartrate
92 c5swcE_
not modelled
27.3
9
PDB header: lyaseChain: E: PDB Molecule: carbonic anhydrase;PDBTitle: the structure of the beta-carbonic anhydrase ccaa
93 c3fiuD_
not modelled
27.1
17
PDB header: ligaseChain: D: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: structure of nmn synthetase from francisella tularensis
94 c2zidA_
not modelled
27.0
12
PDB header: hydrolaseChain: A: PDB Molecule: dextran glucosidase;PDBTitle: crystal structure of dextran glucosidase e236q complex with2 isomaltotriose
95 c3bfjK_
not modelled
26.8
11
PDB header: oxidoreductaseChain: K: PDB Molecule: 1,3-propanediol oxidoreductase;PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
96 c3fkjA_
not modelled
26.5
18
PDB header: isomeraseChain: A: PDB Molecule: putative phosphosugar isomerases;PDBTitle: crystal structure of a putative phosphosugar isomerase (stm_0572) from2 salmonella typhimurium lt2 at 2.12 a resolution
97 d1t57a_
not modelled
26.3
23
Fold: Pyruvate kinase C-terminal domain-likeSuperfamily: PK C-terminal domain-likeFamily: MTH1675-like
98 c4bmvH_
not modelled
26.0
20
PDB header: oxidoreductaseChain: H: PDB Molecule: short-chain dehydrogenase;PDBTitle: short-chain dehydrogenase from sphingobium yanoikuyae in2 complex with nadph
99 c3d3kD_
not modelled
25.2
9
PDB header: protein bindingChain: D: PDB Molecule: enhancer of mrna-decapping protein 3;PDBTitle: crystal structure of human edc3p
100 c3wtcB_
not modelled
24.7
23
PDB header: oxidoreductaseChain: B: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of gox2036
101 c4fn4A_
not modelled
24.6
19
PDB header: oxidoreductaseChain: A: PDB Molecule: short chain dehydrogenase;PDBTitle: short-chain nad(h)-dependent dehydrogenase/reductase from sulfolobus2 acidocaldarius
102 c3ak4C_
not modelled
24.6
12
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-dependent quinuclidinone reductase;PDBTitle: crystal structure of nadh-dependent quinuclidinone reductase from2 agrobacterium tumefaciens
103 d1uoka2
not modelled
24.6
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain
104 c5wqnD_
not modelled
24.5
17
PDB header: oxidoreductaseChain: D: PDB Molecule: probable dehydrogenase;PDBTitle: crystal structure of a carbonyl reductase from pseudomonas aeruginosa2 pao1 (condition ii)
105 d1v71a1
not modelled
24.4
11
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
106 c5ff9C_
not modelled
24.1
15
PDB header: oxidoreductaseChain: C: PDB Molecule: noroxomaritidine/norcraugsodine reductase;PDBTitle: noroxomaritidine/norcraugsodine reductase in complex with nadp+ and2 tyramine
107 d1xq1a_
not modelled
24.0
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
108 c2d1fA_
not modelled
23.9
16
PDB header: lyaseChain: A: PDB Molecule: threonine synthase;PDBTitle: structure of mycobacterium tuberculosis threonine synthase
109 d2hrca1
not modelled
23.8
19
Fold: Chelatase-likeSuperfamily: ChelataseFamily: Ferrochelatase
110 c3pzvB_
not modelled
23.7
12
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase;PDBTitle: c2 crystal form of the endo-1,4-beta-glucanase from bacillus subtilis2 168
111 d1p5ja_
not modelled
23.3
13
Fold: Tryptophan synthase beta subunit-like PLP-dependent enzymesSuperfamily: Tryptophan synthase beta subunit-like PLP-dependent enzymesFamily: Tryptophan synthase beta subunit-like PLP-dependent enzymes
112 c1p5jA_
not modelled
23.3
13
PDB header: lyaseChain: A: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure analysis of human serine dehydratase
113 c4iboA_
not modelled
23.3
17
PDB header: oxidoreductaseChain: A: PDB Molecule: gluconate dehydrogenase;PDBTitle: crystal structure of a putative gluconate dehydrogenase from2 agrobacterium tumefaciens (target efi-506446)
114 c5jc8C_
not modelled
22.6
20
PDB header: oxidoreductaseChain: C: PDB Molecule: putative short-chain dehydrogenase/reductase;PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from burkholderia xenovorans
115 c6gwuB_
not modelled
22.1
13
PDB header: lyaseChain: B: PDB Molecule: carbonic anhydrase;PDBTitle: carbonic anhydrase cance103p from candida albicans
116 c5ak1A_
not modelled
21.8
5
PDB header: hydrolaseChain: A: PDB Molecule: carbohydrate binding family 6;PDBTitle: the penta-modular cellulosomal arabinoxylanase ctxyl5a2 structure as revealed by x-ray crystallography
117 c3amlA_
not modelled
21.4
8
PDB header: transferaseChain: A: PDB Molecule: os06g0726400 protein;PDBTitle: structure of the starch branching enzyme i (bei) from oryza sativa l
118 d1usua_
not modelled
21.4
16
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Hsp90 middle domain
119 c5k9zB_
not modelled
21.4
23
PDB header: oxidoreductaseChain: B: PDB Molecule: putative short-chain dehydrogenase/reductase;PDBTitle: crystal structure of putative short-chain dehydrogenase/reductase from2 burkholderia xenovorans lb400
120 c5zcrB_
not modelled
21.3
13
PDB header: hydrolaseChain: B: PDB Molecule: maltooligosyl trehalose synthase;PDBTitle: dsm5389 glycosyltrehalose synthase