| 1 |
|
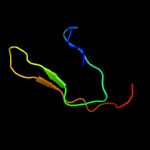
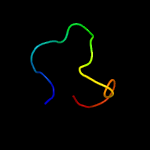
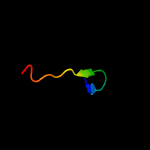
PDB 4b03 chain D
Region: 69 - 104
Aligned: 36
Modelled: 36
Confidence: 70.2%
Identity: 31%
PDB header:virus
Chain: D: PDB Molecule:dengue virus 1 prm protein;
PDBTitle: 6a electron cryomicroscopy structure of immature dengue virus serotype2 1
Phyre2
| 2 |
|
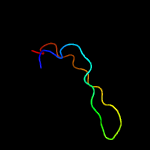
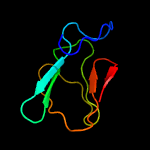
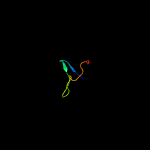
PDB 3c6r chain E
Region: 77 - 104
Aligned: 28
Modelled: 28
Confidence: 61.3%
Identity: 18%
PDB header:virus
Chain: E: PDB Molecule:peptide pr;
PDBTitle: low ph immature dengue virus
Phyre2
| 3 |
|
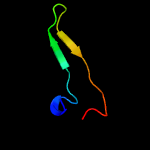
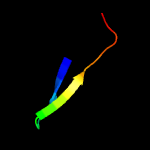
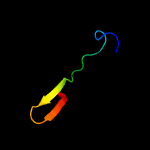
PDB 3c6e chain C
Region: 77 - 104
Aligned: 28
Modelled: 28
Confidence: 61.2%
Identity: 18%
PDB header:viral protein
Chain: C: PDB Molecule:prm;
PDBTitle: crystal structure of the precursor membrane protein- envelope protein2 heterodimer from the dengue 2 virus at neutral ph
Phyre2
| 4 |
|
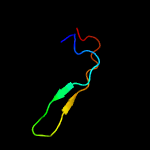
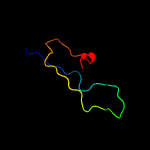
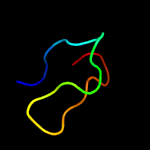
PDB 3ixx chain E
Region: 77 - 104
Aligned: 28
Modelled: 28
Confidence: 56.1%
Identity: 18%
PDB header:virus
Chain: E: PDB Molecule:peptide pr;
PDBTitle: the pseudo-atomic structure of west nile immature virus in complex2 with fab fragments of the anti-fusion loop antibody e53
Phyre2
| 5 |
|
PDB 4edq chain A
Region: 100 - 184
Aligned: 83
Modelled: 85
Confidence: 23.7%
Identity: 24%
PDB header:transport protein/contractile protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein,myosin-binding protein
PDBTitle: mbp-fusion protein of myosin-binding protein c residues 149-269
Phyre2
| 6 |
|
PDB 6rdu chain 9
Region: 113 - 132
Aligned: 20
Modelled: 20
Confidence: 23.6%
Identity: 30%
PDB header:proton transport
Chain: 9: PDB Molecule:asa-9: polytomella f-atp synthase associated subunit 9;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1e,2 monomer-masked refinement
Phyre2
| 7 |
|
PDB 2nt0 chain A domain 1
Region: 83 - 98
Aligned: 16
Modelled: 16
Confidence: 17.6%
Identity: 25%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: Composite domain of glycosyl hydrolase families 5, 30, 39 and 51
Phyre2
| 8 |
|
PDB 3uot chain B
Region: 109 - 173
Aligned: 65
Modelled: 65
Confidence: 12.0%
Identity: 22%
PDB header:cell cycle
Chain: B: PDB Molecule:mediator of dna damage checkpoint protein 1;
PDBTitle: crystal structure of mdc1 fha domain in complex with a phosphorylated2 peptide from the mdc1 n-terminus
Phyre2
| 9 |
|
PDB 5o60 chain W
Region: 128 - 170
Aligned: 43
Modelled: 43
Confidence: 10.9%
Identity: 12%
PDB header:ribosome
Chain: W: PDB Molecule:50s ribosomal protein l25;
PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
Phyre2
| 10 |
|
PDB 5umw chain A
Region: 85 - 102
Aligned: 18
Modelled: 18
Confidence: 10.1%
Identity: 39%
PDB header:tiancimycin-binding protein
Chain: A: PDB Molecule:glyoxalase/bleomycin resisance protein/dioxygenase;
PDBTitle: crystal structure of tnms2, an antibiotic binding protein from2 streptomyces sp. cb03234
Phyre2
| 11 |
|
PDB 2ea9 chain A domain 1
Region: 164 - 178
Aligned: 15
Modelled: 15
Confidence: 9.1%
Identity: 27%
Fold: Profilin-like
Superfamily: YeeU-like
Family: YagB/YeeU/YfjZ-like
Phyre2
| 12 |
|
PDB 1feu chain A
Region: 128 - 170
Aligned: 43
Modelled: 43
Confidence: 8.9%
Identity: 21%
Fold: Ribosomal protein L25-like
Superfamily: Ribosomal protein L25-like
Family: Ribosomal protein L25-like
Phyre2
| 13 |
|
PDB 2h28 chain A domain 1
Region: 164 - 178
Aligned: 15
Modelled: 15
Confidence: 8.2%
Identity: 20%
Fold: Profilin-like
Superfamily: YeeU-like
Family: YagB/YeeU/YfjZ-like
Phyre2
| 14 |
|
PDB 4lqb chain A
Region: 85 - 102
Aligned: 18
Modelled: 18
Confidence: 8.1%
Identity: 11%
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein kfla3161
Phyre2
| 15 |
|
PDB 2inw chain A domain 1
Region: 164 - 178
Aligned: 15
Modelled: 15
Confidence: 7.6%
Identity: 27%
Fold: Profilin-like
Superfamily: YeeU-like
Family: YagB/YeeU/YfjZ-like
Phyre2
| 16 |
|
PDB 6ou9 chain A
Region: 169 - 200
Aligned: 32
Modelled: 32
Confidence: 7.5%
Identity: 19%
PDB header:virus like particle
Chain: A: PDB Molecule:major capsid protein;
PDBTitle: asymmetric focused reconstruction of human norovirus gi.7 houston2 strain vlp asymmetric unit in t=3 symmetry
Phyre2
| 17 |
|
PDB 2kie chain A
Region: 169 - 196
Aligned: 28
Modelled: 28
Confidence: 6.8%
Identity: 18%
PDB header:hydrolase
Chain: A: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: a ph domain within ocrl bridges clathrin mediated membrane2 trafficking to phosphoinositide metabolis
Phyre2
| 18 |
|
PDB 6ngg chain B
Region: 165 - 180
Aligned: 16
Modelled: 16
Confidence: 6.3%
Identity: 50%
PDB header:immune system
Chain: B: PDB Molecule:cd160 antigen;
PDBTitle: crystal structure of human cd160 v58m mutant
Phyre2
| 19 |
|
PDB 2be1 chain A
Region: 73 - 97
Aligned: 25
Modelled: 25
Confidence: 6.1%
Identity: 16%
PDB header:transcription
Chain: A: PDB Molecule:serine/threonine-protein kinase/endoribonuclease ire1;
PDBTitle: structure of the compact lumenal domain of yeast ire1
Phyre2
| 20 |
|
PDB 2ml7 chain A
Region: 91 - 113
Aligned: 23
Modelled: 23
Confidence: 5.7%
Identity: 35%
PDB header:unknown function
Chain: A: PDB Molecule:specific abundant protein 3;
PDBTitle: ginsentides: characterization, structure and application of a new2 class of highly stable cystine knot peptides in ginseng
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|