| 1 |
|
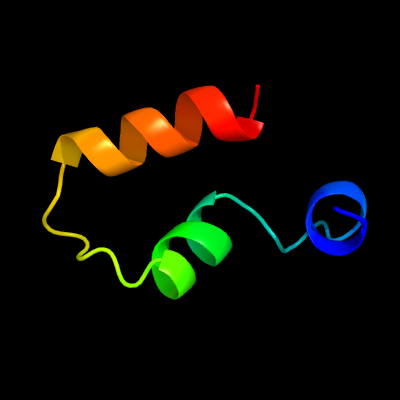
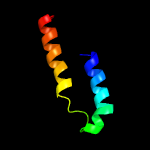
PDB 3tad chain B
Region: 78 - 115
Aligned: 38
Modelled: 38
Confidence: 59.6%
Identity: 24%
PDB header:protein binding
Chain: B: PDB Molecule:liprin-alpha-2;
PDBTitle: crystal structure of the liprin-alpha/liprin-beta complex
Phyre2
| 2 |
|
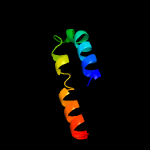
PDB 2e74 chain F domain 1
Region: 25 - 47
Aligned: 23
Modelled: 23
Confidence: 34.4%
Identity: 22%
Fold: Single transmembrane helix
Superfamily: PetM subunit of the cytochrome b6f complex
Family: PetM subunit of the cytochrome b6f complex
Phyre2
| 3 |
|
PDB 2zt9 chain F
Region: 25 - 47
Aligned: 23
Modelled: 23
Confidence: 31.6%
Identity: 22%
PDB header:photosynthesis
Chain: F: PDB Molecule:cytochrome b6-f complex subunit 7;
PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
Phyre2
| 4 |
|
PDB 4h44 chain F
Region: 25 - 47
Aligned: 23
Modelled: 23
Confidence: 31.6%
Identity: 22%
PDB header:photosynthesis
Chain: F: PDB Molecule:cytochrome b6-f complex subunit 7;
PDBTitle: 2.70 a cytochrome b6f complex structure from nostoc pcc 7120
Phyre2
| 5 |
|
PDB 4ogq chain F
Region: 25 - 47
Aligned: 23
Modelled: 23
Confidence: 31.6%
Identity: 22%
PDB header:electron transport
Chain: F: PDB Molecule:cytochrome b6-f complex subunit 7;
PDBTitle: internal lipid architecture of the hetero-oligomeric cytochrome b6f2 complex
Phyre2
| 6 |
|
PDB 2cpb chain A
Region: 21 - 45
Aligned: 25
Modelled: 25
Confidence: 29.2%
Identity: 24%
PDB header:viral protein
Chain: A: PDB Molecule:m13 major coat protein;
PDBTitle: solution nmr structures of the major coat protein of2 filamentous bacteriophage m13 solubilized in3 dodecylphosphocholine micelles, 25 lowest energy structures
Phyre2
| 7 |
|
PDB 3jcu chain E
Region: 74 - 93
Aligned: 20
Modelled: 20
Confidence: 17.9%
Identity: 25%
PDB header:membrane protein
Chain: E: PDB Molecule:cytochrome b559 subunit alpha;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
Phyre2
| 8 |
|
PDB 3kzi chain E
Region: 74 - 93
Aligned: 20
Modelled: 20
Confidence: 16.8%
Identity: 25%
PDB header:electron transport
Chain: E: PDB Molecule:cytochrome b559 subunit alpha;
PDBTitle: crystal structure of monomeric form of cyanobacterial photosystem ii
Phyre2
| 9 |
|
PDB 2axt chain E domain 1
Region: 74 - 93
Aligned: 20
Modelled: 20
Confidence: 15.9%
Identity: 25%
Fold: Single transmembrane helix
Superfamily: Cytochrome b559 subunits
Family: Cytochrome b559 subunits
Phyre2
| 10 |
|
PDB 5wlb chain C
Region: 166 - 175
Aligned: 10
Modelled: 10
Confidence: 12.6%
Identity: 40%
PDB header:protein binding
Chain: C: PDB Molecule:225-15 b;
PDBTitle: kras g12v, bound to gppnhp and miniprotein 225-15a/b
Phyre2
| 11 |
|
PDB 6fki chain B
Region: 87 - 119
Aligned: 33
Modelled: 33
Confidence: 10.1%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:atp synthase subunit beta, chloroplastic;
PDBTitle: chloroplast f1fo conformation 3
Phyre2
| 12 |
|
PDB 5xv8 chain A
Region: 131 - 165
Aligned: 35
Modelled: 35
Confidence: 9.4%
Identity: 14%
PDB header:nuclear protein
Chain: A: PDB Molecule:uv-stimulated scaffold protein a;
PDBTitle: solution structure of the complex between uvssa acidic region and2 tfiih p62 ph domain
Phyre2
| 13 |
|
PDB 5ktf chain A
Region: 73 - 85
Aligned: 13
Modelled: 13
Confidence: 9.3%
Identity: 38%
PDB header:signaling protein
Chain: A: PDB Molecule:scavenger receptor class b member 1;
PDBTitle: structure of the c-terminal transmembrane domain of scavenger receptor2 bi (sr-bi)
Phyre2
| 14 |
|
PDB 6gcs chain G
Region: 87 - 103
Aligned: 17
Modelled: 17
Confidence: 9.3%
Identity: 24%
PDB header:oxidoreductase
Chain: G: PDB Molecule:30-kda protein (nugm);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
Phyre2
| 15 |
|
PDB 4v1g chain A
Region: 14 - 102
Aligned: 79
Modelled: 89
Confidence: 8.3%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:f0f1 atp synthase subunit c;
PDBTitle: crystal structure of a mycobacterial atp synthase rotor ring
Phyre2
| 16 |
|
PDB 5oun chain A
Region: 143 - 163
Aligned: 21
Modelled: 21
Confidence: 6.9%
Identity: 24%
PDB header:hydrolase
Chain: A: PDB Molecule:ruvb-like protein 2;
PDBTitle: nmr solution structure of the external dii domain of rvb2 from2 saccharomyces cerevisiae
Phyre2
| 17 |
|
PDB 2i5n chain H domain 2
Region: 11 - 33
Aligned: 23
Modelled: 23
Confidence: 6.8%
Identity: 17%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 18 |
|
PDB 4k7e chain A
Region: 84 - 95
Aligned: 12
Modelled: 12
Confidence: 6.7%
Identity: 42%
PDB header:viral protein
Chain: A: PDB Molecule:nucleoprotein;
PDBTitle: crystal structure of junin virus nucleoprotein
Phyre2
| 19 |
|
PDB 2mpn chain B
Region: 15 - 58
Aligned: 44
Modelled: 44
Confidence: 5.9%
Identity: 20%
PDB header:membrane protein
Chain: B: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 20 |
|
PDB 2mpn chain A
Region: 15 - 58
Aligned: 44
Modelled: 44
Confidence: 5.9%
Identity: 20%
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein ygap;
PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
Phyre2
| 21 |
|