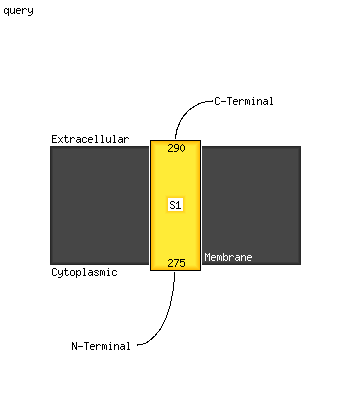
1 c4zhjA_
100.0
28
PDB header: metal binding proteinChain: A: PDB Molecule: mg-chelatase subunit chlh;PDBTitle: crystal structure of the catalytic subunit of magnesium chelatase
2 c3g85A_
88.9
13
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (laci family);PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
3 d2ajta2
88.7
21
Fold: FucI/AraA N-terminal and middle domainsSuperfamily: FucI/AraA N-terminal and middle domainsFamily: AraA N-terminal and middle domain-like
4 c4r1oF_
84.4
16
PDB header: isomeraseChain: F: PDB Molecule: l-arabinose isomerase;PDBTitle: crystal structure of thermophilic geobacillus kaustophilus l-arabinose2 isomerase
5 d2iafa1
77.2
18
Fold: FwdE/GAPDH domain-likeSuperfamily: Serine metabolism enzymes domainFamily: Serine dehydratase beta chain-like
6 c1l9xA_
74.8
17
PDB header: hydrolaseChain: A: PDB Molecule: gamma-glutamyl hydrolase;PDBTitle: structure of gamma-glutamyl hydrolase
7 d1l9xa_
74.8
17
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)
8 c4lqlF_
74.6
13
PDB header: isomeraseChain: F: PDB Molecule: l-arabinose isomerase;PDBTitle: crystal structure of l-arabinose isomerase from lactobacillus2 fermentum cgmcc2921
9 c2f59B_
72.8
15
PDB header: transferaseChain: B: PDB Molecule: 6,7-dimethyl-8-ribityllumazine synthase 1;PDBTitle: lumazine synthase ribh1 from brucella abortus (gene bruab1_0785,2 swiss-prot entry q57dy1) complexed with inhibitor 5-nitro-6-(d-3 ribitylamino)-2,4(1h,3h) pyrimidinedione
10 c4e14A_
72.7
10
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: kynurenine formamidase;PDBTitle: crystal structure of kynurenine formamidase conjugated with2 phenylmethylsulfonyl fluoride
11 c5nn7A_
69.2
20
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: kshv uracil-dna glycosylase, apo form
12 d3euga_
69.2
20
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
13 c3aerC_
69.0
11
PDB header: oxidoreductaseChain: C: PDB Molecule: light-independent protochlorophyllide reductase subunit n;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
14 c4plaA_
68.3
20
PDB header: transferase,hydrolaseChain: A: PDB Molecule: chimera protein of phosphatidylinositol 4-kinase type 2-PDBTitle: crystal structure of phosphatidyl inositol 4-kinase ii alpha in2 complex with atp
15 c4l8fA_
68.0
20
PDB header: hydrolaseChain: A: PDB Molecule: gamma-glutamyl hydrolase;PDBTitle: crystal structure of gamma-glutamyl hydrolase (c108a) complex with mtx
16 d1okba_
67.9
17
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
17 d2hxma1
67.5
15
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
18 c3fijD_
67.1
25
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: lin1909 protein;PDBTitle: crystal structure of a uncharacterized protein lin1909
19 c2booA_
67.0
18
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the crystal structure of uracil-dna n-glycosylase (ung) from2 deinococcus radiodurans.
20 c4ggmX_
66.2
20
PDB header: hydrolaseChain: X: PDB Molecule: udp-2,3-diacylglucosamine pyrophosphatase lpxi;PDBTitle: structure of lpxi
21 c4rqoB_
not modelled
64.4
18
PDB header: lyaseChain: B: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure of l-serine dehydratase from legionella pneumophila
22 c3cxmA_
not modelled
63.6
16
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: leishmania naiffi uracil-dna glycosylase in complex with 5-bromouracil
23 d1jhfa1
not modelled
63.0
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain
24 d2pbla1
not modelled
62.2
11
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Carboxylesterase
25 c3d54D_
not modelled
62.0
17
PDB header: ligaseChain: D: PDB Molecule: phosphoribosylformylglycinamidine synthase 1;PDBTitle: structure of purlqs from thermotoga maritima
26 d1laue_
not modelled
57.5
26
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
27 d1stza1
not modelled
56.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Heat-inducible transcription repressor HrcA, N-terminal domain
28 c5x55A_
not modelled
56.3
14
PDB header: hydrolaseChain: A: PDB Molecule: probable uracil-dna glycosylase;PDBTitle: crystal structure of mimivirus uracil-dna glycosylase
29 c3tr7A_
not modelled
55.8
15
PDB header: hydrolaseChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
30 c2owrD_
not modelled
54.7
17
PDB header: hydrolaseChain: D: PDB Molecule: uracil-dna glycosylase;PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
31 d1uc8a1
not modelled
54.3
19
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Lysine biosynthesis enzyme LysX, N-terminal domain
32 d1m1na_
not modelled
53.9
12
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
33 c4n5iX_
not modelled
53.1
8
PDB header: hydrolaseChain: X: PDB Molecule: esterase/lipase;PDBTitle: crystal structure of a c8-c4 sn3 inhibited esterase b from2 lactobacillus rhamnosis
34 d2j8xa1
not modelled
52.9
21
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Uracil-DNA glycosylase
35 c3zoqA_
not modelled
52.7
16
PDB header: hydrolase/viral proteinChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: structure of bsudg-p56 complex
36 c3okaA_
not modelled
52.6
11
PDB header: transferaseChain: A: PDB Molecule: gdp-mannose-dependent alpha-(1-6)-phosphatidylinositolPDBTitle: crystal structure of corynebacterium glutamicum pimb' in complex with2 gdp-man (triclinic crystal form)
37 c2xdqB_
not modelled
51.6
13
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
38 c3uugB_
not modelled
51.3
15
PDB header: sugar binding proteinChain: B: PDB Molecule: multiple sugar-binding periplasmic receptor chve;PDBTitle: crystal structure of the periplasmic sugar binding protein chve
39 d1qvwa_
not modelled
51.0
24
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: DJ-1/PfpI
40 c3aerB_
not modelled
48.4
15
PDB header: oxidoreductaseChain: B: PDB Molecule: light-independent protochlorophyllide reductase subunit b;PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
41 c4byfA_
not modelled
48.1
25
PDB header: hydrolaseChain: A: PDB Molecule: unconventional myosin-ic;PDBTitle: crystal structure of human myosin 1c in complex with2 calmodulin in the pre-power stroke state
42 d1iowa1
not modelled
46.6
21
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: D-Alanine ligase N-terminal domain
43 c5x3hA_
not modelled
46.0
20
PDB header: dna binding proteinChain: A: PDB Molecule: uracil-dna glycosylase;PDBTitle: the y81g mutant of the ung crystal structure from nitratifractor2 salsuginis
44 c5hsgA_
not modelled
45.1
22
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter, nucleotide binding/atpasePDBTitle: crystal structure of an abc transporter solute binding protein from2 klebsiella pneumoniae (kpn_01730, target efi-511059), apo open3 structure
45 d1qh8a_
not modelled
43.8
14
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
46 d2iw1a1
not modelled
42.4
25
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: Glycosyl transferases group 1
47 c3h3mB_
not modelled
42.4
19
PDB header: structural genomicsChain: B: PDB Molecule: flagellar protein flit;PDBTitle: crystal structure of flagellar protein flit from bordetella2 bronchiseptica
48 c4pd3B_
not modelled
42.1
23
PDB header: contractile proteinChain: B: PDB Molecule: nonmuscle myosin heavy chain b, alpha-actinin a chimeraPDBTitle: crystal structure of rigor-like human nonmuscle myosin-2b
49 c5kp8A_
not modelled
41.7
13
PDB header: transferaseChain: A: PDB Molecule: curd;PDBTitle: crystal structure of the curacin biosynthetic pathway hmg synthase in2 complex with acetyl donor-acp
50 d1q7ra_
not modelled
40.4
21
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)
51 c3gv0A_
not modelled
40.0
12
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator, laci family;PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
52 c1br2C_
not modelled
39.9
21
PDB header: muscle proteinChain: C: PDB Molecule: myosin;PDBTitle: smooth muscle myosin motor domain complexed with mgadp.alf4
53 c5vzmB_
not modelled
39.8
42
PDB header: protein bindingChain: B: PDB Molecule: dna repair protein rev1;PDBTitle: solution nmr structure of human rev1 (932-1039) in complex with2 ubiquitin
54 c3c4vB_
not modelled
39.0
16
PDB header: transferaseChain: B: PDB Molecule: predicted glycosyltransferases;PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
55 d2isya1
not modelled
38.7
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein
56 c3nkzD_
not modelled
38.4
23
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: flagellar protein flit;PDBTitle: the crystal structure of a flagella protein from yersinia2 enterocolitica subsp. enterocolitica 8081
57 c4nq3B_
not modelled
38.0
22
PDB header: hydrolaseChain: B: PDB Molecule: cyanuric acid amidohydrolase;PDBTitle: crystal structure of cyanuic acid hydrolase from a. caulinodans
58 d1g3wa1
not modelled
37.7
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Iron-dependent repressor protein
59 c2qq1A_
not modelled
37.7
21
PDB header: structural proteinChain: A: PDB Molecule: molybdenum cofactor biosynthesis mog;PDBTitle: crystal structure of molybdenum cofactor biosynthesis2 (aq_061) other form from aquifex aeolicus vf5
60 c3kklA_
not modelled
37.1
17
PDB header: hydrolaseChain: A: PDB Molecule: probable chaperone protein hsp33;PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
61 d2f7wa1
not modelled
36.8
17
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like
62 c4x0oG_
not modelled
36.8
12
PDB header: transferaseChain: G: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 2;PDBTitle: beta-ketoacyl-(acyl carrier protein) synthase iii-2 (fabh2) from2 vibrio cholerae soaked with acetyl-coa
63 c3q9tB_
not modelled
36.8
21
PDB header: oxidoreductaseChain: B: PDB Molecule: choline dehydrogenase and related flavoproteins;PDBTitle: crystal structure analysis of formate oxidase
64 d1kk8a2
not modelled
36.4
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
65 c3bjrA_
not modelled
36.3
21
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxylesterase;PDBTitle: crystal structure of a putative carboxylesterase (lp_1002) from2 lactobacillus plantarum wcfs1 at 2.09 a resolution
66 c5tbyA_
not modelled
36.1
21
PDB header: contractile proteinChain: A: PDB Molecule: myosin-7;PDBTitle: human beta cardiac heavy meromyosin interacting-heads motif obtained2 by homology modeling (using swiss-model) of human sequence from3 aphonopelma homology model (pdb-3jbh), rigidly fitted to human beta-4 cardiac negatively stained thick filament 3d-reconstruction (emd-5 2240)
67 c3hxkB_
not modelled
36.0
16
PDB header: hydrolaseChain: B: PDB Molecule: sugar hydrolase;PDBTitle: crystal structure of a sugar hydrolase (yeeb) from lactococcus lactis,2 northeast structural genomics consortium target kr108
68 c4h1hB_
not modelled
35.9
20
PDB header: hydrolaseChain: B: PDB Molecule: lmo1638 protein;PDBTitle: crystal structure of mccf homolog from listeria monocytogenes egd-e
69 c5hw4C_
not modelled
35.6
16
PDB header: transferaseChain: C: PDB Molecule: ribosomal rna small subunit methyltransferase i;PDBTitle: crystal structure of escherichia coli 16s rrna methyltransferase rsmi2 in complex with adomet
70 d1mioa_
not modelled
35.1
13
Fold: Chelatase-likeSuperfamily: "Helical backbone" metal receptorFamily: Nitrogenase iron-molybdenum protein
71 c4l79A_
not modelled
35.1
27
PDB header: motor protein/metal binding proteinChain: A: PDB Molecule: unconventional myosin-ib;PDBTitle: crystal structure of nucleotide-free myosin 1b residues 1-728 with2 bound calmodulin
72 c5tx7A_
not modelled
34.3
27
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 from desulfovibrio vulgaris
73 c3bblA_
not modelled
34.1
12
PDB header: regulatory proteinChain: A: PDB Molecule: regulatory protein of laci family;PDBTitle: crystal structure of a regulatory protein of laci family from2 chloroflexus aggregans
74 c5n6yD_
not modelled
34.1
10
PDB header: oxidoreductaseChain: D: PDB Molecule: nitrogenase vanadium-iron protein alpha chain;PDBTitle: azotobacter vinelandii vanadium nitrogenase
75 d1c41a_
not modelled
33.8
10
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase
76 c1w9iA_
not modelled
33.5
20
PDB header: myosinChain: A: PDB Molecule: myosin ii heavy chain;PDBTitle: myosin ii dictyostelium discoideum motor domain s456y bound2 with mgadp-befx
77 d1j0xo1
not modelled
33.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
78 c3sf5D_
not modelled
33.1
23
PDB header: chaperoneChain: D: PDB Molecule: urease accessory protein ureh;PDBTitle: crystal structure of helicobacter pylori urease accessory protein2 uref/h complex
79 d1lkxa_
not modelled
32.8
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
80 c2fp3A_
not modelled
32.7
12
PDB header: hydrolysis/apoptosisChain: A: PDB Molecule: caspase nc;PDBTitle: crystal structure of the drosophila initiator caspase dronc
81 d2c4va1
not modelled
32.6
5
Fold: Flavodoxin-likeSuperfamily: Type II 3-dehydroquinate dehydrataseFamily: Type II 3-dehydroquinate dehydratase
82 c5a4nB_
not modelled
32.5
5
PDB header: dna binding proteinChain: B: PDB Molecule: bpsl1147;PDBTitle: crystal structure of bpsl1147, a pc4 homolog from burkholderia2 pseudomallei k96243 (tetragonal crystal form)
83 d1d0xa2
not modelled
32.5
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Motor proteins
84 c1x9eB_
not modelled
32.1
19
PDB header: lyaseChain: B: PDB Molecule: hmg-coa synthase;PDBTitle: crystal structure of hmg-coa synthase from enterococcus2 faecalis
85 c3gjzB_
not modelled
31.8
17
PDB header: immune systemChain: B: PDB Molecule: microcin immunity protein mccf;PDBTitle: crystal structure of microcin immunity protein mccf from bacillus2 anthracis str. ames
86 d1tlfa_
not modelled
31.2
17
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like
87 c5kp0A_
not modelled
31.1
25
PDB header: chaperoneChain: A: PDB Molecule: flagellar protein flit,flagellum-specific atp synthase;PDBTitle: recognition and targeting mechanisms by chaperones in flagella2 assembly and operation
88 c3gg9C_
not modelled
31.1
24
PDB header: oxidoreductaseChain: C: PDB Molecule: d-3-phosphoglycerate dehydrogenase oxidoreductase protein;PDBTitle: crystal structure of putative d-3-phosphoglycerate dehydrogenase2 oxidoreductase from ralstonia solanacearum
89 c4xkjA_
not modelled
30.9
21
PDB header: oxidoreductaseChain: A: PDB Molecule: d-lactate dehydrogenase;PDBTitle: a novel d-lactate dehydrogenase from sporolactobacillus sp
90 c3pdiG_
not modelled
30.1
21
PDB header: protein bindingChain: G: PDB Molecule: nitrogenase mofe cofactor biosynthesis protein nife;PDBTitle: precursor bound nifen
91 c5dteD_
not modelled
30.0
12
PDB header: transport proteinChain: D: PDB Molecule: monosaccharide-transporting atpase;PDBTitle: crystal structure of an abc transporter periplasmic solute binding2 protein (ipr025997) from actinobacillus succinogenes 130z(asuc_0081,3 target efi-511065) with bound d-allose
92 c6ch2E_
not modelled
30.0
26
PDB header: structural proteinChain: E: PDB Molecule: flagellar hook-associated protein 2,flagellar protein flit;PDBTitle: crystal structure of the cytoplasmic domain of flha and flit-flid2 complex
93 d1nqua_
not modelled
29.9
20
Fold: Lumazine synthaseSuperfamily: Lumazine synthaseFamily: Lumazine synthase
94 d2a5la1
not modelled
29.8
27
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like
95 c5wb4H_
not modelled
29.8
14
PDB header: transferaseChain: H: PDB Molecule: n-acetylglucosaminyldiphosphoundecaprenol n-acetyl-beta-d-PDBTitle: crystal structure of the tara wall teichoic acid glycosyltransferase
96 d1u8fo1
not modelled
29.6
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
97 c2zkiH_
not modelled
29.5
16
PDB header: transcriptionChain: H: PDB Molecule: 199aa long hypothetical trp repressor bindingPDBTitle: crystal structure of hypothetical trp repressor binding2 protein from sul folobus tokodaii (st0872)
98 c4rxuA_
not modelled
28.9
8
PDB header: transport proteinChain: A: PDB Molecule: periplasmic sugar-binding protein;PDBTitle: crystal structure of carbohydrate transporter solute binding protein2 caur_1924 from chloroflexus aurantiacus, target efi-511158, in3 complex with d-glucose
99 d2dria_
not modelled
28.9
14
Fold: Periplasmic binding protein-like ISuperfamily: Periplasmic binding protein-like IFamily: L-arabinose binding protein-like
100 d1hdgo1
not modelled
28.9
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
101 c2q62A_
not modelled
28.7
13
PDB header: flavoproteinChain: A: PDB Molecule: arsh;PDBTitle: crystal structure of arsh from sinorhizobium meliloti
102 d1t92a_
not modelled
28.5
20
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pseudopilin
103 c3av0A_
not modelled
28.2
11
PDB header: recombinationChain: A: PDB Molecule: dna double-strand break repair protein mre11;PDBTitle: crystal structure of mre11-rad50 bound to atp s
104 c2kwuA_
not modelled
27.9
38
PDB header: protein binding/signaling proteinChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
105 c4jenB_
not modelled
27.9
14
PDB header: hydrolaseChain: B: PDB Molecule: cmp n-glycosidase;PDBTitle: structure of clostridium botulinum cmp n-glycosidase, bcmb
106 c5tkaA_
not modelled
27.8
12
PDB header: metal binding proteinChain: A: PDB Molecule: oxsa protein;PDBTitle: structure of the hd-domain phosphohydrolase oxsa
107 d1t3ta2
not modelled
27.7
17
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)
108 c3ahyD_
not modelled
27.5
18
PDB header: hydrolaseChain: D: PDB Molecule: beta-glucosidase;PDBTitle: crystal structure of beta-glucosidase 2 from fungus trichoderma reesei2 in complex with tris
109 c3jbhA_
not modelled
27.0
20
PDB header: contractile proteinChain: A: PDB Molecule: myosin 2 heavy chain striated muscle;PDBTitle: two heavy meromyosin interacting-heads motifs flexible docked into2 tarantula thick filament 3d-map allows in depth study of intra- and3 intermolecular interactions
110 d2paqa1
not modelled
26.9
20
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain
111 c3il3A_
not modelled
26.8
12
PDB header: transferaseChain: A: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] synthase 3;PDBTitle: structure of haemophilus influenzae fabh
112 c3sipC_
not modelled
26.6
22
PDB header: hydrolase/ligase/hydrolaseChain: C: PDB Molecule: caspase;PDBTitle: crystal structure of drice and diap1-bir1 complex
113 c3h5oB_
not modelled
26.6
16
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator gntr;PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
114 d1qfma2
not modelled
26.4
10
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Prolyl oligopeptidase, C-terminal domain
115 d1ggaa1
not modelled
26.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
116 c3huuC_
not modelled
26.3
13
PDB header: transcription regulatorChain: C: PDB Molecule: transcription regulator like protein;PDBTitle: crystal structure of transcription regulator like protein from2 staphylococcus haemolyticus
117 c2kepA_
not modelled
26.0
15
PDB header: transport proteinChain: A: PDB Molecule: general secretion pathway protein g;PDBTitle: solution structure of xcpt, the main component of the type 22 secretion system of pseudomonas aeruginosa
118 c4fl0A_
not modelled
26.0
12
PDB header: transferaseChain: A: PDB Molecule: aminotransferase ald1;PDBTitle: crystal structure of ald1 from arabidopsis thaliana
119 d1gado1
not modelled
25.8
30
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
120 c4gxhC_
not modelled
25.5
19
PDB header: hydrolaseChain: C: PDB Molecule: pyrrolidone-carboxylate peptidase;PDBTitle: crystal structure of a pyrrolidone-carboxylate peptidase 1 (target id2 nysgrc-012831) from xenorhabdus bovienii ss-2004