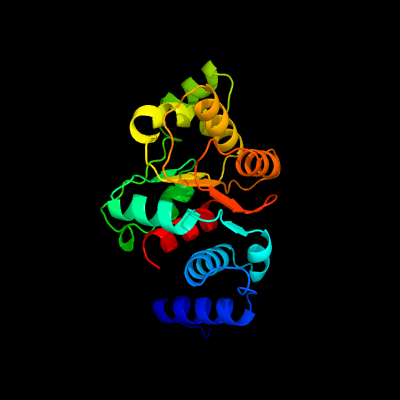
| 1 | d1f2va_
|
|
 |
100.0 |
55 |
Fold:Flavodoxin-like
Superfamily:Precorrin-8X methylmutase CbiC/CobH
Family:Precorrin-8X methylmutase CbiC/CobH |
|
|
|
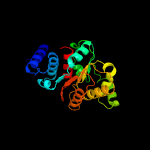
| 2 | c3e7dC_
|
|
 |
100.0 |
57 |
PDB header:isomerase
Chain: C: PDB Molecule:cobh, precorrin-8x methylmutase;
PDBTitle: crystal structure of precorrin-8x methyl mutase cbic/cobh from2 brucella melitensis
|
|
|
|
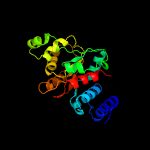
| 3 | c4au1A_
|
|
 |
100.0 |
51 |
PDB header:isomerase
Chain: A: PDB Molecule:precorrin-8x methylmutase;
PDBTitle: crystal structure of cobh (precorrin-8x methyl mutase)2 complexed with c5 desmethyl-hba
|
|
|
|
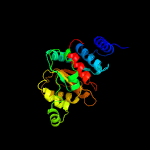
| 4 | c2afvB_
|
|
 |
100.0 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin isomerase;
PDBTitle: the crystal structure of putative precorrin isomerase cbic2 in cobalamin biosynthesis
|
|
|
|
| 5 | d1ou0a_
|
|
 |
100.0 |
34 |
Fold:Flavodoxin-like
Superfamily:Precorrin-8X methylmutase CbiC/CobH
Family:Precorrin-8X methylmutase CbiC/CobH |
|
|
|
| 6 | d1v9ca_
|
|
 |
100.0 |
36 |
Fold:Flavodoxin-like
Superfamily:Precorrin-8X methylmutase CbiC/CobH
Family:Precorrin-8X methylmutase CbiC/CobH |
|
|
|
| 7 | d1miob_
|
|
 |
88.5 |
9 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
|
| 8 | c5kojD_
|
|
 |
88.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nitrogenase femo beta subunit protein nifk;
PDBTitle: nitrogenase mofep protein in the ids oxidized state
|
|
|
|
| 9 | d2ji7a1
|
|
 |
85.9 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 10 | d1ovma1
|
|
 |
85.9 |
12 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 11 | d1ybha1
|
|
 |
83.5 |
21 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 12 | d1zpda1
|
|
 |
82.3 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 13 | d1ozha1
|
|
 |
82.0 |
17 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 14 | d2djia1
|
|
 |
79.9 |
19 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 15 | d1pvda1
|
|
 |
79.8 |
16 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 16 | d1qh8b_
|
|
 |
78.0 |
14 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
|
| 17 | d2ihta1
|
|
 |
74.6 |
23 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 18 | c3aerB_
|
|
 |
74.4 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the light-independent protochlorophyllide reductase2 catalyzing a key reduction for greening in the dark
|
|
|
|
| 19 | d2ez9a1
|
|
 |
70.6 |
18 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
|
| 20 | d1m1nb_
|
|
 |
68.6 |
16 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
|
| 21 | c2ynmD_ |
|
not modelled |
66.9 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: structure of the adpxalf3-stabilized transition state of the2 nitrogenase-like dark-operative protochlorophyllide oxidoreductase3 complex from prochlorococcus marinus with its substrate4 protochlorophyllide a
|
|
|
| 22 | d1t9ba1 |
|
not modelled |
65.7 |
15 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 23 | c3cf4G_ |
|
not modelled |
58.1 |
4 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:acetyl-coa decarboxylase/synthase epsilon subunit;
PDBTitle: structure of the codh component of the m. barkeri acds complex
|
|
|
| 24 | c5n6yE_ |
|
not modelled |
56.2 |
14 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nitrogenase vanadium-iron protein beta chain;
PDBTitle: azotobacter vinelandii vanadium nitrogenase
|
|
|
| 25 | d1q6za1 |
|
not modelled |
49.5 |
22 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Pyruvate oxidase and decarboxylase, middle domain |
|
|
| 26 | c2rnjA_ |
|
not modelled |
41.4 |
8 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein vrar;
PDBTitle: nmr structure of the s. aureus vrar dna binding domain
|
|
|
| 27 | d3bofa1 |
|
not modelled |
39.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Methyltetrahydrofolate-utiluzing methyltransferases |
|
|
| 28 | d1qh8a_ |
|
not modelled |
38.9 |
21 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 29 | c4grdA_ |
|
not modelled |
37.8 |
23 |
PDB header:lyase,isomerase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from burkholderia cenocepacia j2315
|
|
|
| 30 | c4wzzA_ |
|
not modelled |
37.8 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:putative sugar abc transporter, substrate-binding protein;
PDBTitle: crystal structure of an abc transporter solute binding protein2 (ipr025997) from clostridium phytofermentas (cphy_0583, target efi-3 511148) with bound l-rhamnose
|
|
|
| 31 | d1mioa_ |
|
not modelled |
36.5 |
27 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 32 | d1jcea1 |
|
not modelled |
36.3 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 33 | c1zljE_ |
|
not modelled |
34.6 |
11 |
PDB header:transcription
Chain: E: PDB Molecule:dormancy survival regulator;
PDBTitle: crystal structure of the mycobacterium tuberculosis hypoxic2 response regulator dosr c-terminal domain
|
|
|
| 34 | c4rjjB_ |
|
not modelled |
34.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:acetolactate synthase;
PDBTitle: acetolactate synthase from bacillus subtilis bound to thdp - crystal2 form ii
|
|
|
| 35 | c2xdqB_ |
|
not modelled |
34.2 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:light-independent protochlorophyllide reductase subunit b;
PDBTitle: dark operative protochlorophyllide oxidoreductase (chln-chlb)2 complex
|
|
|
| 36 | d1m1na_ |
|
not modelled |
34.1 |
18 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 37 | d1a9xa2 |
|
not modelled |
34.1 |
16 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
|
|
| 38 | c1zpdA_ |
|
not modelled |
32.2 |
13 |
PDB header:alcohol fermentation
Chain: A: PDB Molecule:pyruvate decarboxylase;
PDBTitle: pyruvate decarboxylase from zymomonas mobilis
|
|
|
| 39 | c1ozhD_ |
|
not modelled |
31.6 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:acetolactate synthase, catabolic;
PDBTitle: the crystal structure of klebsiella pneumoniae acetolactate2 synthase with enzyme-bound cofactor and with an unusual3 intermediate.
|
|
|
| 40 | c2krfB_ |
|
not modelled |
30.8 |
5 |
PDB header:transcription
Chain: B: PDB Molecule:transcriptional regulatory protein coma;
PDBTitle: nmr solution structure of the dna binding domain of competence protein2 a
|
|
|
| 41 | c2vbgB_ |
|
not modelled |
30.5 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:branched-chain alpha-ketoacid decarboxylase;
PDBTitle: the complex structure of the branched-chain keto acid2 decarboxylase (kdca) from lactococcus lactis with 2r-1-3 hydroxyethyl-deazathdp
|
|
|
| 42 | c1powA_ |
|
not modelled |
29.8 |
20 |
PDB header:oxidoreductase(oxygen as acceptor)
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: the refined structures of a stabilized mutant and of wild-type2 pyruvate oxidase from lactobacillus plantarum
|
|
|
| 43 | c6o55B_ |
|
not modelled |
28.9 |
25 |
PDB header:isomerase
Chain: B: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 (pure) from legionella pneumophila
|
|
|
| 44 | d1a04a1 |
|
not modelled |
28.3 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
|
|
| 45 | c2ji6B_ |
|
not modelled |
28.2 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:oxalyl-coa decarboxylase;
PDBTitle: x-ray structure of oxalyl-coa decarboxylase in complex with 3-deaza-2 thdp and oxalyl-coa
|
|
|
| 46 | c1ovmC_ |
|
not modelled |
27.7 |
11 |
PDB header:lyase
Chain: C: PDB Molecule:indole-3-pyruvate decarboxylase;
PDBTitle: crystal structure of indolepyruvate decarboxylase from2 enterobacter cloacae
|
|
|
| 47 | c5z2fA_ |
|
not modelled |
27.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
|
|
|
| 48 | d1p4wa_ |
|
not modelled |
27.0 |
5 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
|
|
| 49 | c4ww4B_ |
|
not modelled |
26.3 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:ruvb-like 2;
PDBTitle: double-heterohexameric rings of full-length rvb1(adp)/rvb2(adp)
|
|
|
| 50 | c3sftA_ |
|
not modelled |
25.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:chemotaxis response regulator protein-glutamate
PDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
|
|
|
| 51 | d1o4va_ |
|
not modelled |
25.4 |
20 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 52 | c4ja0A_ |
|
not modelled |
25.2 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase;
PDBTitle: crystal structure of the invertebrate bi-functional purine2 biosynthesis enzyme paics at 2.8 a resolution
|
|
|
| 53 | c2nxwB_ |
|
not modelled |
25.2 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:phenyl-3-pyruvate decarboxylase;
PDBTitle: crystal structure of phenylpyruvate decarboxylase of azospirillum2 brasilense
|
|
|
| 54 | c3pdiB_ |
|
not modelled |
24.3 |
24 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
|
|
| 55 | c3ib7A_ |
|
not modelled |
23.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:icc protein;
PDBTitle: crystal structure of full length rv0805
|
|
|
| 56 | c5tenH_ |
|
not modelled |
23.6 |
20 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:4-hydroxy-tetrahydrodipicolinate reductase;
PDBTitle: structure of 4-hydroxy-tetrahydrodipicolinate reductase from vibrio2 vulnificus with 2,5 furan dicarboxylic and nadh with intact3 polyhistidine tag
|
|
|
| 57 | d1l3la1 |
|
not modelled |
23.6 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:C-terminal effector domain of the bipartite response regulators
Family:GerE-like (LuxR/UhpA family of transcriptional regulators) |
|
|
| 58 | c2fw9A_ |
|
not modelled |
23.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: structure of pure (n5-carboxyaminoimidazole ribonucleotide mutase)2 h59f from the acidophilic bacterium acetobacter aceti, at ph 8
|
|
|
| 59 | c2vbiF_ |
|
not modelled |
22.8 |
19 |
PDB header:lyase
Chain: F: PDB Molecule:pyruvate decarboxylase;
PDBTitle: holostructure of pyruvate decarboxylase from acetobacter pasteurianus
|
|
|
| 60 | c2w93A_ |
|
not modelled |
22.8 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:pyruvate decarboxylase isozyme 1;
PDBTitle: crystal structure of the saccharomyces cerevisiae pyruvate2 decarboxylase variant e477q in complex with the surrogate pyruvamide
|
|
|
| 61 | d1u11a_ |
|
not modelled |
22.7 |
23 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 62 | c2h31A_ |
|
not modelled |
21.5 |
20 |
PDB header:ligase, lyase
Chain: A: PDB Molecule:multifunctional protein ade2;
PDBTitle: crystal structure of human paics, a bifunctional carboxylase and2 synthetase in purine biosynthesis
|
|
|
| 63 | d2fsja2 |
|
not modelled |
20.6 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ta0583-like |
|
|
| 64 | c2yvqA_ |
|
not modelled |
20.6 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
|
|
| 65 | c6gcsK_ |
|
not modelled |
20.6 |
24 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:psst subunit (nukm);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
| 66 | c2panF_ |
|
not modelled |
20.4 |
17 |
PDB header:lyase
Chain: F: PDB Molecule:glyoxylate carboligase;
PDBTitle: crystal structure of e. coli glyoxylate carboligase
|
|
|
| 67 | d1d7ya1 |
|
not modelled |
20.3 |
20 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 68 | d2hk6a1 |
|
not modelled |
20.1 |
20 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
|
|
| 69 | c3rggD_ |
|
not modelled |
19.6 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
|
|
|
| 70 | c1yi1A_ |
|
not modelled |
18.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase;
PDBTitle: crystal structure of arabidopsis thaliana acetohydroxyacid synthase in2 complex with a sulfonylurea herbicide, tribenuron methyl
|
|
|
| 71 | c2pgnA_ |
|
not modelled |
18.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:cyclohexane-1,2-dione hydrolase (cdh);
PDBTitle: the crystal structure of fad and thdp-dependent cyclohexane-1,2-dione2 hydrolase in complex with cyclohexane-1,2-dione
|
|
|
| 72 | c1upaC_ |
|
not modelled |
18.1 |
23 |
PDB header:synthase
Chain: C: PDB Molecule:carboxyethylarginine synthase;
PDBTitle: carboxyethylarginine synthase from streptomyces2 clavuligerus (semet structure)
|
|
|
| 73 | c2djiA_ |
|
not modelled |
18.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate oxidase;
PDBTitle: crystal structure of pyruvate oxidase from aerococcus2 viridans containing fad
|
|
|
| 74 | c4e21B_ |
|
not modelled |
18.1 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:6-phosphogluconate dehydrogenase (decarboxylating);
PDBTitle: the crystal structure of 6-phosphogluconate dehydrogenase from2 geobacter metallireducens
|
|
|
| 75 | d1chda_ |
|
not modelled |
18.0 |
21 |
Fold:Methylesterase CheB, C-terminal domain
Superfamily:Methylesterase CheB, C-terminal domain
Family:Methylesterase CheB, C-terminal domain |
|
|
| 76 | c3sztB_ |
|
not modelled |
17.8 |
6 |
PDB header:transcription
Chain: B: PDB Molecule:quorum-sensing control repressor;
PDBTitle: quorum sensing control repressor, qscr, bound to n-3-oxo-dodecanoyl-l-2 homoserine lactone
|
|
|
| 77 | c3sajB_ |
|
not modelled |
17.8 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor 1;
PDBTitle: crystal structure of glutamate receptor glua1 amino terminal domain
|
|
|
| 78 | c2jzfA_ |
|
not modelled |
17.8 |
23 |
PDB header:viral protein
Chain: A: PDB Molecule:replicase polyprotein 1ab;
PDBTitle: nmr conformer closest to the mean coordinates of the domain 513-651 of2 the sars-cov nonstructural protein nsp3
|
|
|
| 79 | c4oaqA_ |
|
not modelled |
16.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:r-specific carbonyl reductase;
PDBTitle: crystal structure of the r-specific carbonyl reductase from candida2 parapsilosis atcc 7330
|
|
|
| 80 | c6humK_ |
|
not modelled |
16.7 |
24 |
PDB header:proton transport
Chain: K: PDB Molecule:nad(p)h-quinone oxidoreductase subunit k;
PDBTitle: structure of the photosynthetic complex i from thermosynechococcus2 elongatus
|
|
|
| 81 | c3q3hA_ |
|
not modelled |
16.4 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:hmw1c-like glycosyltransferase;
PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
|
|
|
| 82 | c4b4kK_ |
|
not modelled |
16.3 |
21 |
PDB header:isomerase
Chain: K: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of bacillus anthracis pure
|
|
|
| 83 | d1xmpa_ |
|
not modelled |
16.0 |
21 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 84 | d2hrca1 |
|
not modelled |
15.5 |
15 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:Ferrochelatase |
|
|
| 85 | c6ioxB_ |
|
not modelled |
15.2 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:phosphotransacetylase;
PDBTitle: crystal structure of porphyromonas gingivalis phosphotransacetylase in2 complex with acetyl-coa
|
|
|
| 86 | d1zfja1 |
|
not modelled |
14.7 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Inosine monophosphate dehydrogenase (IMPDH)
Family:Inosine monophosphate dehydrogenase (IMPDH) |
|
|
| 87 | c5yw2D_ |
|
not modelled |
14.6 |
16 |
PDB header:transferase
Chain: D: PDB Molecule:adenine phosphoribosyltransferase;
PDBTitle: crystal structure of adenine phosphoribosyltransferase from2 francisella tularensis.
|
|
|
| 88 | d1qcza_ |
|
not modelled |
14.3 |
25 |
Fold:Flavodoxin-like
Superfamily:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE)
Family:N5-CAIR mutase (phosphoribosylaminoimidazole carboxylase, PurE) |
|
|
| 89 | c4cpdA_ |
|
not modelled |
14.1 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase tadh from thermus sp. atn1
|
|
|
| 90 | c2kqvA_ |
|
not modelled |
13.9 |
16 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 3;
PDBTitle: sars coronavirus-unique domain (sud): three-domain molecular2 architecture in solution and rna binding. i: structure of the sud-m3 domain of sud-mc
|
|
|
| 91 | c4lfuA_ |
|
not modelled |
13.5 |
6 |
PDB header:dna binding protein
Chain: A: PDB Molecule:regulatory protein sdia;
PDBTitle: crystal structure of escherichia coli sdia in the space group c2
|
|
|
| 92 | d1k4ia_ |
|
not modelled |
13.5 |
16 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
|
|
| 93 | d1zcha1 |
|
not modelled |
13.4 |
28 |
Fold:FMN-dependent nitroreductase-like
Superfamily:FMN-dependent nitroreductase-like
Family:NADH oxidase/flavin reductase |
|
|
| 94 | d1vm6a3 |
|
not modelled |
13.4 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 95 | c3lp6D_ |
|
not modelled |
13.4 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of rv3275c-e60a from mycobacterium tuberculosis at2 1.7a resolution
|
|
|
| 96 | c4ffjA_ |
|
not modelled |
13.3 |
33 |
PDB header:lyase
Chain: A: PDB Molecule:riboflavin biosynthesis protein ribba;
PDBTitle: the crystal structure of spdhbps from s.pneumoniae
|
|
|
| 97 | c1m6vE_ |
|
not modelled |
13.3 |
17 |
PDB header:ligase
Chain: E: PDB Molecule:carbamoyl phosphate synthetase large chain;
PDBTitle: crystal structure of the g359f (small subunit) point mutant of2 carbamoyl phosphate synthetase
|
|
|
| 98 | d3bzka5 |
|
not modelled |
13.2 |
26 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
|
|
| 99 | d1snna_ |
|
not modelled |
13.2 |
33 |
Fold:YrdC/RibB
Superfamily:YrdC/RibB
Family:3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB |
|
|