| 1 | c3nutC_
|
|
 |
100.0 |
44 |
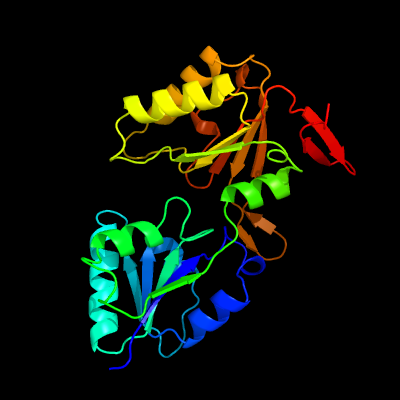
PDB header:transferase
Chain: C: PDB Molecule:precorrin-3 methylase;
PDBTitle: crystal structure of the methyltransferase cobj
|
|
|
|
| 2 | c2zvbA_
|
|
 |
100.0 |
40 |
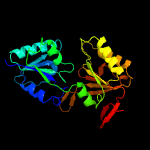
PDB header:transferase
Chain: A: PDB Molecule:precorrin-3 c17-methyltransferase;
PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
|
|
|
|
| 3 | c2e0kA_
|
|
 |
100.0 |
29 |
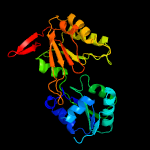
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
|
|
|
| 4 | c1pjtB_
|
|
 |
100.0 |
22 |
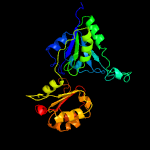
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
|
| 5 | c2yboA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
|
|
|
| 6 | c2qbuA_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
|
|
|
| 7 | d1s4da_
|
|
 |
100.0 |
20 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 8 | d1cbfa_
|
|
 |
100.0 |
20 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 9 | c1cbfA_
|
|
 |
100.0 |
20 |
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
|
|
|
| 10 | d1pjqa2
|
|
 |
100.0 |
23 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 11 | c3kwpA_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
|
| 12 | c3ndcB_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
|
|
|
| 13 | c4e16A_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: precorrin-4 c(11)-methyltransferase from clostridium difficile
|
|
|
|
| 14 | d1ve2a1
|
|
 |
100.0 |
26 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 15 | d1va0a1
|
|
 |
100.0 |
26 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 16 | c5hw4C_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:ribosomal rna small subunit methyltransferase i;
PDBTitle: crystal structure of escherichia coli 16s rrna methyltransferase rsmi2 in complex with adomet
|
|
|
|
| 17 | c3nd1B_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
|
|
|
| 18 | d1wyza1
|
|
 |
100.0 |
13 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 19 | c2npnA_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
|
|
|
| 20 | d2deka1
|
|
 |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 21 | c5n0sA_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
| 22 | d1vhva_ |
|
not modelled |
100.0 |
15 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 23 | c3i4tA_ |
|
not modelled |
100.0 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diphthine synthase;
PDBTitle: crystal structure of putative diphthine synthase from entamoeba2 histolytica
|
|
|
| 24 | d1wdea_ |
|
not modelled |
100.0 |
25 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 25 | c2bb3B_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
|
|
| 26 | d2bb3a1 |
|
not modelled |
100.0 |
25 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 27 | c3hh1D_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:tetrapyrrole methylase family protein;
PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
|
|
|
| 28 | c3fq6A_ |
|
not modelled |
98.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
|
|
| 29 | c3vndD_ |
|
not modelled |
93.6 |
16 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 30 | c5ey5A_ |
|
not modelled |
90.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 31 | c3navB_ |
|
not modelled |
88.7 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 32 | c5kzmA_ |
|
not modelled |
88.3 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 33 | c2ekcA_ |
|
not modelled |
79.4 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 34 | d1qopa_ |
|
not modelled |
72.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 35 | d1q7sa_ |
|
not modelled |
72.0 |
28 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 36 | c2d3kA_ |
|
not modelled |
70.2 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: structural study on project id ph1539 from pyrococcus2 horikoshii ot3
|
|
|
| 37 | d3erja1 |
|
not modelled |
63.5 |
21 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 38 | c4bucA_ |
|
not modelled |
58.7 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
|
|
|
| 39 | c3zq4C_ |
|
not modelled |
56.5 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:ribonuclease j 1;
PDBTitle: unusual, dual endo- and exo-nuclease activity in the degradosome2 explained by crystal structure analysis of rnase j1
|
|
|
| 40 | c3fpjA_ |
|
not modelled |
56.1 |
13 |
PDB header:biosynthetic protein, transferase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
|
|
|
| 41 | c5k9xA_ |
|
not modelled |
55.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
| 42 | c1xtyD_ |
|
not modelled |
54.6 |
20 |
PDB header:hydrolase
Chain: D: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of sulfolobus solfataricus peptidyl-trna hydrolase
|
|
|
| 43 | c6c4nB_ |
|
not modelled |
53.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pseudopaline dehydrogenase;
PDBTitle: pseudopaline dehydrogenase (paodh) - nadp+ bound
|
|
|
| 44 | c3b1fA_ |
|
not modelled |
51.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative prephenate dehydrogenase;
PDBTitle: crystal structure of prephenate dehydrogenase from streptococcus2 mutans
|
|
|
| 45 | d2p1ra1 |
|
not modelled |
50.8 |
18 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 46 | c3k30B_ |
|
not modelled |
46.9 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: histamine dehydrogenase from nocardiodes simplex
|
|
|
| 47 | d2jfga2 |
|
not modelled |
46.1 |
15 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
| 48 | d1rlka_ |
|
not modelled |
44.9 |
21 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 49 | c2a5hC_ |
|
not modelled |
43.2 |
19 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 50 | c4txkA_ |
|
not modelled |
40.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein-methionine sulfoxide oxidase mical1;
PDBTitle: construct of mical-1 containing the monooxygenase and calponin2 homology domains
|
|
|
| 51 | d2o8ra3 |
|
not modelled |
40.2 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 52 | c2zv3E_ |
|
not modelled |
40.2 |
25 |
PDB header:hydrolase
Chain: E: PDB Molecule:peptidyl-trna hydrolase;
PDBTitle: crystal structure of project mj0051 from methanocaldococcus2 jannaschii dsm 2661
|
|
|
| 53 | c5th5C_ |
|
not modelled |
38.3 |
7 |
PDB header:lyase
Chain: C: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
|
|
|
| 54 | d2hzba1 |
|
not modelled |
37.6 |
14 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
|
|
| 55 | c5zbyA_ |
|
not modelled |
36.6 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrogenase maturation protease hyci;
PDBTitle: crystal structure of a [nife] hydrogenase maturation protease hyci2 from thermococcus kodakarensis kod1
|
|
|
| 56 | c3mvnA_ |
|
not modelled |
35.9 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-medo-
PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamyl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
|
|
|
| 57 | c2q7xA_ |
|
not modelled |
35.8 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:upf0052 protein sp_1565;
PDBTitle: crystal structure of a putative phospho transferase (sp_1565) from2 streptococcus pneumoniae tigr4 at 2.00 a resolution
|
|
|
| 58 | c5jg7A_ |
|
not modelled |
33.2 |
20 |
PDB header:metal transport
Chain: A: PDB Molecule:fur regulated salmonella iron transporter;
PDBTitle: crystal structure of putative periplasmic binding protein from2 salmonella typhimurium lt2
|
|
|
| 59 | c3l7oB_ |
|
not modelled |
32.8 |
27 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-phosphate isomerase a from streptococcus2 mutans ua159
|
|
|
| 60 | d1cfza_ |
|
not modelled |
32.7 |
17 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Hydrogenase maturating endopeptidase HybD |
|
|
| 61 | c2ppvA_ |
|
not modelled |
32.4 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
|
|
| 62 | c1lkzB_ |
|
not modelled |
31.4 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase a;
PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia) from2 escherichia coli.
|
|
|
| 63 | c3rp7A_ |
|
not modelled |
31.1 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavoprotein monooxygenase;
PDBTitle: crystal structure of klebsiella pneumoniae hpxo complexed with fad and2 uric acid
|
|
|
| 64 | c6c4rA_ |
|
not modelled |
30.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:staphylopine dehydrogenase;
PDBTitle: staphylopine dehydrogenase (saodh) - apo
|
|
|
| 65 | c4xwwA_ |
|
not modelled |
30.2 |
18 |
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:dr2417;
PDBTitle: crystal structure of rnase j complexed with rna
|
|
|
| 66 | d2bona1 |
|
not modelled |
29.0 |
17 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 67 | c5v1tA_ |
|
not modelled |
28.8 |
12 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam;
PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
|
|
|
| 68 | c3lk7A_ |
|
not modelled |
28.5 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-glutamate2 (murd) ligase from streptococcus agalactiae to 1.5a
|
|
|
| 69 | c4mjsQ_ |
|
not modelled |
27.1 |
25 |
PDB header:transferase/protein binding
Chain: Q: PDB Molecule:protein kinase c zeta type;
PDBTitle: crystal structure of a pb1 complex
|
|
|
| 70 | c1vqwB_ |
|
not modelled |
26.6 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein with similarity to flavin-containing
PDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
|
|
|
| 71 | d2qv7a1 |
|
not modelled |
26.3 |
20 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 72 | c3en0A_ |
|
not modelled |
25.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:cyanophycinase;
PDBTitle: the structure of cyanophycinase
|
|
|
| 73 | c2r6r1_ |
|
not modelled |
25.4 |
13 |
PDB header:cell cycle
Chain: 1: PDB Molecule:cell division protein ftsz;
PDBTitle: aquifex aeolicus ftsz
|
|
|
| 74 | c6c8vA_ |
|
not modelled |
25.1 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme pqq synthesis protein e;
PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
|
|
|
| 75 | d1ve5a1 |
|
not modelled |
25.0 |
13 |
Fold:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Superfamily:Tryptophan synthase beta subunit-like PLP-dependent enzymes
Family:Tryptophan synthase beta subunit-like PLP-dependent enzymes |
|
|
| 76 | c3qfeB_ |
|
not modelled |
24.2 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
|
|
| 77 | c1w59B_ |
|
not modelled |
23.2 |
16 |
PDB header:cell division
Chain: B: PDB Molecule:cell division protein ftsz homolog 1;
PDBTitle: ftsz dimer, empty (m. jannaschii)
|
|
|
| 78 | c4d9gA_ |
|
not modelled |
23.1 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:putative diaminopropionate ammonia-lyase;
PDBTitle: crystal structure of selenomethionine incorporated holo2 diaminopropionate ammonia lyase from escherichia coli
|
|
|
| 79 | d1wu2a3 |
|
not modelled |
22.2 |
13 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 80 | c5u57B_ |
|
not modelled |
21.5 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:(s)-2-hydroxypropylphosphonic acid epoxidase;
PDBTitle: psf4 in complex with fe2+ and (s)-2-hpp
|
|
|
| 81 | c3ktsA_ |
|
not modelled |
21.2 |
11 |
PDB header:transcriptional regulator
Chain: A: PDB Molecule:glycerol uptake operon antiterminator regulatory protein;
PDBTitle: crystal structure of glycerol uptake operon antiterminator regulatory2 protein from listeria monocytogenes str. 4b f2365
|
|
|
| 82 | c2bonB_ |
|
not modelled |
21.1 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:lipid kinase;
PDBTitle: structure of an escherichia coli lipid kinase (yegs)
|
|
|
| 83 | d2vapa1 |
|
not modelled |
20.8 |
16 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 84 | c2o8rA_ |
|
not modelled |
20.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
|
|
|
| 85 | d1xdpa3 |
|
not modelled |
20.5 |
15 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 86 | c5t8xA_ |
|
not modelled |
20.3 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prephenate dehydrogenase 1;
PDBTitle: prephenate dehydrogenase from soybean
|
|
|
| 87 | c1xdoB_ |
|
not modelled |
20.2 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
|
|
| 88 | c2e1mA_ |
|
not modelled |
20.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-glutamate oxidase;
PDBTitle: crystal structure of l-glutamate oxidase from streptomyces sp. x-119-6
|
|
|
| 89 | d2bfdb2 |
|
not modelled |
19.7 |
28 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
|
|
| 90 | d1pjqa1 |
|
not modelled |
19.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 91 | c3kwmC_ |
|
not modelled |
19.3 |
15 |
PDB header:isomerase
Chain: C: PDB Molecule:ribose-5-phosphate isomerase a;
PDBTitle: crystal structure of ribose-5-isomerase a
|
|
|
| 92 | c5dboA_ |
|
not modelled |
19.2 |
9 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
|
|
|
| 93 | d1ps9a3 |
|
not modelled |
18.7 |
18 |
Fold:Nucleotide-binding domain
Superfamily:Nucleotide-binding domain
Family:N-terminal domain of adrenodoxin reductase-like |
|
|
| 94 | c3vzdB_ |
|
not modelled |
18.6 |
16 |
PDB header:transferase/inhibitor
Chain: B: PDB Molecule:sphingosine kinase 1;
PDBTitle: crystal structure of sphingosine kinase 1 with inhibitor and adp
|
|
|
| 95 | c3h14A_ |
|
not modelled |
18.6 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:aminotransferase, classes i and ii;
PDBTitle: crystal structure of a putative aminotransferase from silicibacter2 pomeroyi
|
|
|
| 96 | c2qv7A_ |
|
not modelled |
18.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:diacylglycerol kinase dgkb;
PDBTitle: crystal structure of diacylglycerol kinase dgkb in complex with adp2 and mg
|
|
|
| 97 | d2ffea1 |
|
not modelled |
18.4 |
13 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
|
|
| 98 | d1kyqa1 |
|
not modelled |
18.4 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 99 | c1fncA_ |
|
not modelled |
18.3 |
9 |
PDB header:oxidoreductase (nadp+(a),ferredoxin(a))
Chain: A: PDB Molecule:ferredoxin-nadp+ reductase;
PDBTitle: refined crystal structure of spinach ferredoxin reductase2 at 1.7 angstroms resolution: oxidized, reduced, and 2'-3 phospho-5'-amp bound states
|
|
|