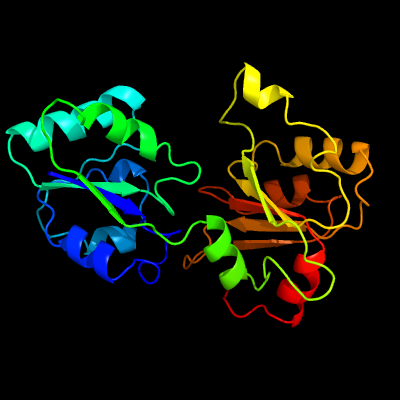
| 1 | c1pjtB_
|
|
 |
100.0 |
28 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
|
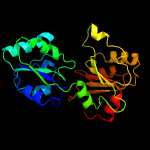
| 2 | c1cbfA_
|
|
 |
100.0 |
40 |
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
|
|
|
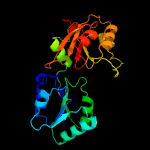
| 3 | d1cbfa_
|
|
 |
100.0 |
40 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
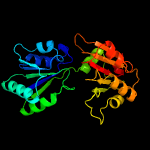
| 4 | d1s4da_
|
|
 |
100.0 |
29 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 5 | c2yboA_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
|
|
|
| 6 | c3ndcB_
|
|
 |
100.0 |
55 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
|
|
|
| 7 | c4e16A_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: precorrin-4 c(11)-methyltransferase from clostridium difficile
|
|
|
|
| 8 | d1pjqa2
|
|
 |
100.0 |
28 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 9 | c3kwpA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
|
| 10 | d1ve2a1
|
|
 |
100.0 |
30 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 11 | d1va0a1
|
|
 |
100.0 |
28 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 12 | c5hw4C_
|
|
 |
100.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:ribosomal rna small subunit methyltransferase i;
PDBTitle: crystal structure of escherichia coli 16s rrna methyltransferase rsmi2 in complex with adomet
|
|
|
|
| 13 | c2zvbA_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-3 c17-methyltransferase;
PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
|
|
|
|
| 14 | d2deka1
|
|
 |
100.0 |
20 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 15 | d1vhva_
|
|
 |
100.0 |
15 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 16 | d1wdea_
|
|
 |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 17 | c3nutC_
|
|
 |
100.0 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:precorrin-3 methylase;
PDBTitle: crystal structure of the methyltransferase cobj
|
|
|
|
| 18 | d1wyza1
|
|
 |
100.0 |
10 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 19 | c3i4tA_
|
|
 |
100.0 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diphthine synthase;
PDBTitle: crystal structure of putative diphthine synthase from entamoeba2 histolytica
|
|
|
|
| 20 | c2e0kA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
|
|
|
| 21 | c2qbuA_ |
|
not modelled |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
|
|
| 22 | c2npnA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
|
|
| 23 | c5n0sA_ |
|
not modelled |
100.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
| 24 | c3nd1B_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
|
|
| 25 | c2bb3B_ |
|
not modelled |
100.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
|
|
| 26 | d2bb3a1 |
|
not modelled |
100.0 |
18 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 27 | c3hh1D_ |
|
not modelled |
99.9 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:tetrapyrrole methylase family protein;
PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
|
|
|
| 28 | c3fq6A_ |
|
not modelled |
99.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
|
|
| 29 | d2o8ra3 |
|
not modelled |
88.4 |
14 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 30 | c2o8rA_ |
|
not modelled |
82.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
|
|
|
| 31 | c5v1tA_ |
|
not modelled |
74.6 |
18 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam;
PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
|
|
|
| 32 | d2p1ra1 |
|
not modelled |
68.4 |
13 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 33 | c5th5C_ |
|
not modelled |
67.5 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
|
|
|
| 34 | c6efnA_ |
|
not modelled |
64.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sporulation killing factor maturation protein skfb;
PDBTitle: structure of a ripp maturase, skfb
|
|
|
| 35 | d1j5pa4 |
|
not modelled |
64.1 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 36 | c5er0D_ |
|
not modelled |
63.1 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:nadh oxidase;
PDBTitle: water-forming nadh oxidase from lactobacillus brevis (lbnox)
|
|
|
| 37 | c4njkA_ |
|
not modelled |
59.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
|
|
|
| 38 | d1p3y1_ |
|
not modelled |
59.2 |
9 |
Fold:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Superfamily:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD
Family:Homo-oligomeric flavin-containing Cys decarboxylases, HFCD |
|
|
| 39 | d7reqa2 |
|
not modelled |
58.8 |
10 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 40 | c3mcuF_ |
|
not modelled |
58.7 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dipicolinate synthase, b chain;
PDBTitle: crystal structure of the dipicolinate synthase chain b from bacillus2 cereus. northeast structural genomics consortium target bcr215.
|
|
|
| 41 | c5tueB_ |
|
not modelled |
58.2 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:tetracycline destructase tet(50);
PDBTitle: crystal structure of tetracycline destructase tet(50)
|
|
|
| 42 | c3bioB_ |
|
not modelled |
58.0 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase (gfo/idh/moca family member) from2 porphyromonas gingivalis w83
|
|
|
| 43 | c2dc1A_ |
|
not modelled |
57.3 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-aspartate dehydrogenase;
PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
|
|
|
| 44 | d2jgra1 |
|
not modelled |
56.6 |
13 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 45 | c3lqkA_ |
|
not modelled |
55.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit b;
PDBTitle: crystal structure of dipicolinate synthase subunit b from bacillus2 halodurans c
|
|
|
| 46 | c2cduB_ |
|
not modelled |
54.1 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph oxidase;
PDBTitle: the crystal structure of water-forming nad(p)h oxidase from2 lactobacillus sanfranciscensis
|
|
|
| 47 | c3ic5A_ |
|
not modelled |
52.7 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
|
|
| 48 | c1xdoB_ |
|
not modelled |
52.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of escherichia coli polyphosphate kinase
|
|
|
| 49 | c5dboA_ |
|
not modelled |
51.4 |
15 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of the tetrameric eif2b-beta2-delta2 complex from c.2 thermophilum
|
|
|
| 50 | c6p2iA_ |
|
not modelled |
51.3 |
7 |
PDB header:oxidoreductase, biosynthetic protein
Chain: A: PDB Molecule:glycerate dehydrogenase;
PDBTitle: acyclic imino acid reductase (bsp5) in complex with nadph and d-arg
|
|
|
| 51 | c4bucA_ |
|
not modelled |
51.1 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
|
|
|
| 52 | c6cluC_ |
|
not modelled |
51.0 |
15 |
PDB header:antimicrobial protein
Chain: C: PDB Molecule:dihydropteroate synthase;
PDBTitle: staphylococcus aureus dihydropteroate synthase (sadhps) f17l e208k2 double mutant structure
|
|
|
| 53 | c3oc4A_ |
|
not modelled |
50.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, pyridine nucleotide-disulfide family;
PDBTitle: crystal structure of a pyridine nucleotide-disulfide family2 oxidoreductase from the enterococcus faecalis v583
|
|
|
| 54 | d1yxya1 |
|
not modelled |
49.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 55 | c3wb9A_ |
|
not modelled |
48.5 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diaminopimelate dehydrogenase;
PDBTitle: crystal structures of meso-diaminopimelate dehydrogenase from2 symbiobacterium thermophilum
|
|
|
| 56 | c6a4tB_ |
|
not modelled |
48.3 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:peptidase e;
PDBTitle: crystal structure of peptidase e from deinococcus radiodurans r1
|
|
|
| 57 | c1nhqA_ |
|
not modelled |
47.8 |
19 |
PDB header:oxidoreductase (h2o2(a))
Chain: A: PDB Molecule:nadh peroxidase;
PDBTitle: crystallographic analyses of nadh peroxidase cys42ala and cys42ser2 mutants: active site structure, mechanistic implications, and an3 unusual environment of arg303
|
|
|
| 58 | c5tchG_ |
|
not modelled |
46.9 |
21 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 59 | d1lssa_ |
|
not modelled |
42.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Potassium channel NAD-binding domain |
|
|
| 60 | d2jfga2 |
|
not modelled |
42.7 |
27 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
| 61 | c2a5hC_ |
|
not modelled |
40.6 |
25 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 62 | d1xdpa3 |
|
not modelled |
40.3 |
16 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 63 | c2z2vA_ |
|
not modelled |
40.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical protein ph1688;
PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
|
|
|
| 64 | c4txkA_ |
|
not modelled |
40.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein-methionine sulfoxide oxidase mical1;
PDBTitle: construct of mical-1 containing the monooxygenase and calponin2 homology domains
|
|
|
| 65 | c2ppvA_ |
|
not modelled |
39.4 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
|
|
| 66 | c5jg7A_ |
|
not modelled |
38.5 |
20 |
PDB header:metal transport
Chain: A: PDB Molecule:fur regulated salmonella iron transporter;
PDBTitle: crystal structure of putative periplasmic binding protein from2 salmonella typhimurium lt2
|
|
|
| 67 | c4u0pB_ |
|
not modelled |
38.5 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:lipoyl synthase 2;
PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
|
|
|
| 68 | c2yx0A_ |
|
not modelled |
35.8 |
19 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
|
|
| 69 | c3kpgA_ |
|
not modelled |
35.8 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfide-quinone reductase, putative;
PDBTitle: crystal structure of sulfide:quinone oxidoreductase from2 acidithiobacillus ferrooxidans in complex with decylubiquinone
|
|
|
| 70 | c1u1iC_ |
|
not modelled |
35.7 |
32 |
PDB header:isomerase
Chain: C: PDB Molecule:myo-inositol-1-phosphate synthase;
PDBTitle: myo-inositol phosphate synthase mips from a. fulgidus
|
|
|
| 71 | c6c8vA_ |
|
not modelled |
35.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme pqq synthesis protein e;
PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
|
|
|
| 72 | c3lk7A_ |
|
not modelled |
35.2 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanine--d-glutamate ligase;
PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-glutamate2 (murd) ligase from streptococcus agalactiae to 1.5a
|
|
|
| 73 | d1u1ia1 |
|
not modelled |
34.9 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 74 | c1j5pA_ |
|
not modelled |
34.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aspartate dehydrogenase;
PDBTitle: crystal structure of aspartate dehydrogenase (tm1643) from thermotoga2 maritima at 1.9 a resolution
|
|
|
| 75 | c5b04G_ |
|
not modelled |
34.5 |
17 |
PDB header:translation
Chain: G: PDB Molecule:probable translation initiation factor eif-2b subunit
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 76 | d2jfga1 |
|
not modelled |
33.3 |
21 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 77 | d2q4qa1 |
|
not modelled |
33.2 |
11 |
Fold:MTH938-like
Superfamily:MTH938-like
Family:MTH938-like |
|
|
| 78 | c2z2uA_ |
|
not modelled |
33.1 |
27 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
|
|
| 79 | d1umdb2 |
|
not modelled |
32.7 |
20 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
|
|
| 80 | c3kd9B_ |
|
not modelled |
32.6 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:coenzyme a disulfide reductase;
PDBTitle: crystal structure of pyridine nucleotide disulfide oxidoreductase from2 pyrococcus horikoshii
|
|
|
| 81 | d2ffea1 |
|
not modelled |
32.3 |
14 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
|
|
| 82 | d2ozlb2 |
|
not modelled |
31.7 |
17 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
|
|
| 83 | c5j7xA_ |
|
not modelled |
31.6 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dimethylaniline monooxygenase, putative;
PDBTitle: baeyer-villiger monooxygenase bvmoafl838 from aspergillus flavus
|
|
|
| 84 | d1k0ia1 |
|
not modelled |
31.6 |
26 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 85 | d1f0ka_ |
|
not modelled |
30.8 |
12 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
|
|
| 86 | c6b4cH_ |
|
not modelled |
30.4 |
20 |
PDB header:antiviral protein
Chain: H: PDB Molecule:viperin;
PDBTitle: structure of viperin from trichoderma virens
|
|
|
| 87 | c2bcpA_ |
|
not modelled |
30.3 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase;
PDBTitle: structural analysis of streptococcus pyogenes nadh oxidase: c44s nox2 with azide
|
|
|
| 88 | d1iuka_ |
|
not modelled |
29.0 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:CoA-binding domain |
|
|
| 89 | d2voua1 |
|
not modelled |
28.9 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|
| 90 | c5z2fA_ |
|
not modelled |
28.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dihydrodipicolinate reductase;
PDBTitle: nadph/pda bound dihydrodipicolinate reductase from paenisporosarcina2 sp. tg-14
|
|
|
| 91 | c2bryA_ |
|
not modelled |
28.7 |
27 |
PDB header:transport
Chain: A: PDB Molecule:nedd9 interacting protein with calponin homology
PDBTitle: crystal structure of the native monooxygenase domain of2 mical at 1.45 a resolution
|
|
|
| 92 | c3rhtB_ |
|
not modelled |
27.4 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:(gatase1)-like protein;
PDBTitle: crystal structure of type 1 glutamine amidotransferase (gatase1)-like2 protein from planctomyces limnophilus
|
|
|
| 93 | c1gqqA_ |
|
not modelled |
27.1 |
11 |
PDB header:cell wall biosynthesis
Chain: A: PDB Molecule:udp-n-acetylmuramate-l-alanine ligase;
PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
|
|
|
| 94 | c4s1vD_ |
|
not modelled |
26.9 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:d-3-phosphoglycerate dehydrogenase-related protein;
PDBTitle: crystal structure of phosphoglycerate oxidoreductase from vibrio2 cholerae o395
|
|
|
| 95 | c3uclA_ |
|
not modelled |
26.4 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cyclohexanone monooxygenase;
PDBTitle: cyclohexanone-bound crystal structure of cyclohexanone monooxygenase2 in the rotated conformation
|
|
|
| 96 | c3cgdB_ |
|
not modelled |
26.3 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyridine nucleotide-disulfide oxidoreductase, class i;
PDBTitle: pyridine nucleotide complexes with bacillus anthracis coenzyme a-2 disulfide reductase: a structural analysis of dual nad(p)h3 specificity
|
|
|
| 97 | c5b3uB_ |
|
not modelled |
26.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 98 | c3zidB_ |
|
not modelled |
25.9 |
11 |
PDB header:gtp-binding protein
Chain: B: PDB Molecule:tubulin/ftsz, gtpase;
PDBTitle: cetz from methanosaeta thermophila strain dsm 6194
|
|
|
| 99 | c6bz5B_ |
|
not modelled |
25.8 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:salicylate hydroxylase;
PDBTitle: structure and mechanism of salicylate hydroxylase from pseudomonas2 putida g7
|
|
|
| 100 | c2gr2A_ |
|
not modelled |
25.6 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin reductase;
PDBTitle: crystal structure of ferredoxin reductase, bpha4 (oxidized form)
|
|
|
| 101 | c5do7B_ |
|
not modelled |
25.4 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 8;
PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
|
|
|
| 102 | c2p0yA_ |
|
not modelled |
25.3 |
0 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus plantarum.2 northeast structural genomics consortium target lpr6
|
|
|
| 103 | d2naca2 |
|
not modelled |
25.0 |
9 |
Fold:Flavodoxin-like
Superfamily:Formate/glycerate dehydrogenase catalytic domain-like
Family:Formate/glycerate dehydrogenases, substrate-binding domain |
|
|
| 104 | c5b04B_ |
|
not modelled |
24.9 |
18 |
PDB header:translation
Chain: B: PDB Molecule:translation initiation factor eif-2b subunit alpha;
PDBTitle: crystal structure of the eukaryotic translation initiation factor 2b2 from schizosaccharomyces pombe
|
|
|
| 105 | c4zeoH_ |
|
not modelled |
24.1 |
15 |
PDB header:translation
Chain: H: PDB Molecule:translation initiation factor eif-2b-like protein;
PDBTitle: crystal structure of eif2b delta from chaetomium thermophilum
|
|
|
| 106 | c1vqwB_ |
|
not modelled |
23.9 |
37 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein with similarity to flavin-containing
PDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
|
|
|
| 107 | c3h8lA_ |
|
not modelled |
23.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidase;
PDBTitle: the first x-ray structure of a sulfide:quinone2 oxidoreductase: insights into sulfide oxidation mechanism
|
|
|
| 108 | c2bonB_ |
|
not modelled |
23.4 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:lipid kinase;
PDBTitle: structure of an escherichia coli lipid kinase (yegs)
|
|
|
| 109 | c1ir6A_ |
|
not modelled |
23.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:exonuclease recj;
PDBTitle: crystal structure of exonuclease recj bound to manganese
|
|
|
| 110 | d1ir6a_ |
|
not modelled |
23.0 |
22 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Exonuclease RecJ |
|
|
| 111 | d1w85b2 |
|
not modelled |
22.5 |
26 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain |
|
|
| 112 | c4gmfD_ |
|
not modelled |
22.4 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:yersiniabactin biosynthetic protein ybtu;
PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
|
|
|
| 113 | d2bona1 |
|
not modelled |
22.3 |
14 |
Fold:NAD kinase/diacylglycerol kinase-like
Superfamily:NAD kinase/diacylglycerol kinase-like
Family:Diacylglycerol kinase-like |
|
|
| 114 | d1gpua3 |
|
not modelled |
22.1 |
23 |
Fold:TK C-terminal domain-like
Superfamily:TK C-terminal domain-like
Family:Transketolase C-terminal domain-like |
|
|
| 115 | d2gv8a1 |
|
not modelled |
22.0 |
33 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD/NAD-linked reductases, N-terminal and central domains |
|
|
| 116 | d1ydwa1 |
|
not modelled |
21.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 117 | d1a9xa4 |
|
not modelled |
21.8 |
19 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 118 | c3ezyB_ |
|
not modelled |
21.5 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of probable dehydrogenase tm_0414 from thermotoga2 maritima
|
|
|
| 119 | c5jciA_ |
|
not modelled |
21.2 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:os09g0567300 protein;
PDBTitle: structure and catalytic mechanism of monodehydroascorbate reductase,2 mdhar, from oryza sativa l. japonica
|
|
|
| 120 | c2axqA_ |
|
not modelled |
21.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
|
|