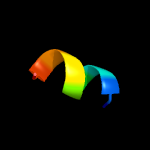
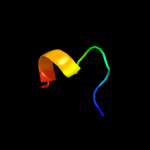
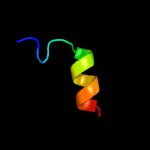
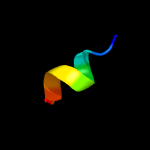
1 c3fwbB_
70.6
31
PDB header: cell cycle, transcriptionChain: B: PDB Molecule: nuclear mrna export protein sac3;PDBTitle: sac3:sus1:cdc31 complex
2 c4ndlC_
38.9
30
PDB header: de novo proteinChain: C: PDB Molecule: enh-c2b, computational designed homodimer;PDBTitle: computational design and experimental verification of a symmetric2 homodimer
3 d2axtz1
34.8
21
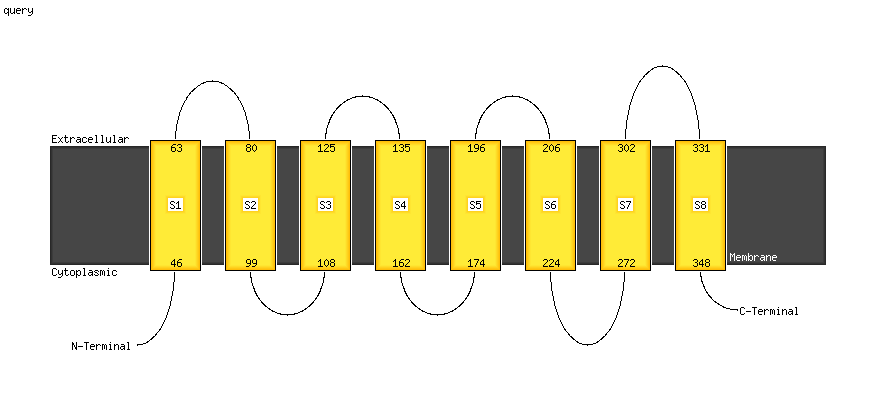
Fold: Transmembrane helix hairpinSuperfamily: PsbZ-likeFamily: PsbZ-like
4 d2obba1
24.0
33
Fold: HAD-likeSuperfamily: HAD-likeFamily: BT0820-like
5 c4ndkA_
23.4
33
PDB header: fluorescent protein, de novo proteinChain: A: PDB Molecule: e23p-yfp, gfp-like fluorescent chromoprotein fp506,PDBTitle: crystal structure of a computational designed engrailed homeodomain2 variant fused with yfp
6 d1b33n_
19.7
36
Fold: Allophycocyanin linker chain (domain)Superfamily: Allophycocyanin linker chain (domain)Family: Allophycocyanin linker chain (domain)
7 d1dw0a_
16.8
19
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
8 d1u6ra1
14.4
42
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
9 d1gu2a_
14.3
31
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c
10 d1m15a1
12.9
50
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
11 d1v8ca2
12.9
56
Fold: TBP-likeSuperfamily: MoaD-related protein, C-terminal domainFamily: MoaD-related protein, C-terminal domain
12 c5lqwL_
12.8
50
PDB header: splicingChain: L: PDB Molecule: pre-mrna-splicing factor cwc26;PDBTitle: yeast activated spliceosome
13 d1vrpa1
12.4
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
14 d1u6ka1
12.1
48
Fold: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)Superfamily: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)Family: F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
15 c5y0tD_
11.4
38
PDB header: ligaseChain: D: PDB Molecule: thermotoga maritima tmcal;PDBTitle: crystal structure of thermotoga maritima tmcal bound with alpha-thio2 atp(form ii)
16 d1g0wa1
11.2
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
17 d1i0ea1
11.0
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
18 d1crka1
10.9
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
19 d1qh4a1
10.3
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
20 d1qk1a1
10.2
40
Fold: Guanido kinase N-terminal domainSuperfamily: Guanido kinase N-terminal domainFamily: Guanido kinase N-terminal domain
21 d1oo2a_
not modelled
9.7
6
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)
22 d1ttaa_
not modelled
9.2
13
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)
23 d1f86a_
not modelled
8.5
12
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)
24 c5ixiB_
not modelled
8.3
54
PDB header: cytokineChain: B: PDB Molecule: chimera protein of interferon lambda receptor 1 andPDBTitle: structure of human jak1 ferm/sh2 in complex with ifnlr1/il10ra chimera
25 d1kgia_
not modelled
7.8
13
Fold: Prealbumin-likeSuperfamily: Transthyretin (synonym: prealbumin)Family: Transthyretin (synonym: prealbumin)
26 c5y0nB_
not modelled
7.6
41
PDB header: ligaseChain: B: PDB Molecule: upf0348 protein b4417_3650;PDBTitle: crystal structure of bacillus subtilis tmcal bound with atp (semet2 derivative)
27 c2dg7A_
not modelled
7.2
23
PDB header: gene regulationChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of the putative transcriptional regulator sco03372 from streptomyces coelicolor a3(2)
28 c6gctA_
not modelled
7.1
16
PDB header: membrane proteinChain: A: PDB Molecule: neutral amino acid transporter b(0);PDBTitle: cryo-em structure of the human neutral amino acid transporter asct2
29 d1hf2a2
not modelled
6.8
16
Fold: Cell-division inhibitor MinC, N-terminal domainSuperfamily: Cell-division inhibitor MinC, N-terminal domainFamily: Cell-division inhibitor MinC, N-terminal domain
30 c3qvaB_
not modelled
6.6
41
PDB header: hydrolaseChain: B: PDB Molecule: transthyretin-like protein;PDBTitle: structure of klebsiella pneumoniae 5-hydroxyisourate hydrolase
31 c2l1nA_
not modelled
6.5
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of the protein yp_399305.1
32 c5vkqC_
not modelled
6.5
17
PDB header: membrane proteinChain: C: PDB Molecule: no mechanoreceptor potential c isoform l;PDBTitle: structure of a mechanotransduction ion channel drosophila nompc in2 nanodisc
33 c2l3wA_
not modelled
6.2
27
PDB header: photosynthesisChain: A: PDB Molecule: phycobilisome rod linker polypeptide;PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome rod2 linker polypeptide from synechococcus elongatus, northeast structural3 genomics consortium target snr168a
34 c4dh4A_
not modelled
6.2
13
PDB header: isomeraseChain: A: PDB Molecule: mif;PDBTitle: macrophage migration inhibitory factor toxoplasma gondii
35 c4z1mJ_
not modelled
6.1
44
PDB header: hydrolaseChain: J: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: bovine f1-atpase inhibited by three copies of the inhibitor protein2 if1 crystallised in the presence of thiophosphate.
36 c2qkwA_
not modelled
6.1
63
PDB header: transferaseChain: A: PDB Molecule: avirulence protein;PDBTitle: structural basis for activation of plant immunity by2 bacterial effector protein avrpto
37 d2qkwa1
not modelled
6.1
63
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Avirulence protein AvrPtoFamily: Avirulence protein AvrPto
38 c4z1mI_
not modelled
6.0
40
PDB header: hydrolaseChain: I: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: bovine f1-atpase inhibited by three copies of the inhibitor protein2 if1 crystallised in the presence of thiophosphate.
39 c4tt3I_
not modelled
6.0
40
PDB header: hydrolaseChain: I: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: the pathway of binding of the intrinsically disordered mitochondrial2 inhibitor protein to f1-atpase
40 c3t5vE_
not modelled
6.0
19
PDB header: transcriptionChain: E: PDB Molecule: nuclear mrna export protein thp1;PDBTitle: sac3:thp1:sem1 complex
41 c5ietA_
not modelled
6.0
19
PDB header: gene regulationChain: A: PDB Molecule: bacterial proteasome activator;PDBTitle: crystal structure of mycobacterium tuberculosis atp-independent2 proteasome activator
42 c2h1xB_
not modelled
6.0
12
PDB header: hydrolaseChain: B: PDB Molecule: 5-hydroxyisourate hydrolase (formerly known asPDBTitle: crystal structure of 5-hydroxyisourate hydrolase (formerly2 known as trp, transthyretin related protein)
43 c3jcuz_
not modelled
5.9
34
PDB header: membrane proteinChain: Z: PDB Molecule: photosystem ii reaction center protein z;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
44 c1ohhH_
not modelled
5.7
40
PDB header: synthaseChain: H: PDB Molecule: atpase inhibitor, mitochondrial;PDBTitle: bovine mitochondrial f1-atpase complexed with the inhibitor2 protein if1
45 d1smye_
not modelled
5.5
38
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RNA polymerase omega subunit
46 c2cw1A_
not modelled
5.5
60
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
47 c3zbeA_
not modelled
5.5
8
PDB header: toxin-antitoxinChain: A: PDB Molecule: paaa2;PDBTitle: e. coli o157 pare2-associated antitoxin 2 (paaa2)
48 c2rukA_
not modelled
5.4
36
PDB header: transcriptionChain: A: PDB Molecule: cellular tumor antigen p53;PDBTitle: solution structure of the complex between p53 transactivation domain 22 and tfiih p62 ph domain
49 d1x4ta1
not modelled
5.4
41
Fold: Long alpha-hairpinSuperfamily: ISY1 domain-likeFamily: ISY1 N-terminal domain-like
50 c1pnbA_
not modelled
5.3
8
PDB header: seed storage proteinChain: A: PDB Molecule: napin bnib;PDBTitle: structure of napin bnib, nmr, 10 structures
51 c1q90M_
not modelled
5.2
33
PDB header: photosynthesisChain: M: PDB Molecule: cytochrome b6f complex subunit petm;PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii