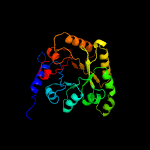
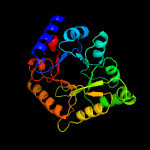
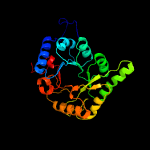
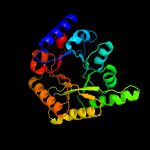
1 c3cpgA_
100.0
34
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an unknown protein from bifidobacterium2 adolescentis
2 c1w8gA_
100.0
33
PDB header: plp-binding proteinChain: A: PDB Molecule: hypothetical upf0001 protein yggs;PDBTitle: crystal structure of e. coli k-12 yggs
3 c3r79B_
100.0
31
PDB header: structure genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharactertized protein from agrobacterium2 tumefaciens
4 d1ct5a_
100.0
28
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: "Hypothetical" protein ybl036c
5 c5nm8A_
100.0
35
PDB header: plp-binding proteinChain: A: PDB Molecule: pipy;PDBTitle: structure of pipy, the cog0325 family member of synechococcus2 elongatus pcc7942, with plp bound
6 c4y2wA_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 1;PDBTitle: crystal structure of a thermostable alanine racemase from2 thermoanaerobacter tengcongensis mb4
7 c4bf5A_
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: structure of broad spectrum racemase from aeromonas hydrophila
8 c3oo2A_
100.0
14
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
9 d1bd0a2
100.0
15
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
10 c4fs9B_
100.0
20
PDB header: isomeraseChain: B: PDB Molecule: broad specificity amino acid racemase;PDBTitle: complex structure of a broad specificity amino acid racemase (bar)2 within the reactive intermediate
11 d1vfsa2
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
12 c2dy3B_
100.0
15
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from corynebacterium glutamicum
13 c5zl6A_
100.0
17
PDB header: isomeraseChain: A: PDB Molecule: histidine racemase;PDBTitle: histidine racemase from leuconostoc mesenteroides subsp. sake nbrc2 102480
14 c3e6eC_
100.0
16
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from e.faecalis2 complex with cycloserine
15 c4eclA_
100.0
14
PDB header: isomeraseChain: A: PDB Molecule: serine racemase;PDBTitle: crystal structure of the cytoplasmic domain of vancomycin resistance2 serine racemase vantg
16 c4beqA_
100.0
18
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 2;PDBTitle: structure of vibrio cholerae broad spectrum racemase double2 mutant r173a, n174a
17 c3mubB_
100.0
15
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: the crystal structure of alanine racemase from streptococcus2 pneumoniae
18 c5irpA_
100.0
13
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase 2;PDBTitle: crystal structure of the alanine racemase bsu17640 from bacillus2 subtilis
19 c1xfcB_
100.0
25
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: the 1.9 a crystal structure of alanine racemase from mycobacterium2 tuberculosis contains a conserved entryway into the active site
20 c5yycC_
100.0
17
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from bacillus pseudofirmus (of4)
21 c3oo2B_
not modelled
100.0
13
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase 1;PDBTitle: 2.37 angstrom resolution crystal structure of an alanine racemase2 (alr) from staphylococcus aureus subsp. aureus col
22 c1niuA_
not modelled
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: alanine racemase with bound inhibitor derived from l-2 cycloserine
23 c3co8B_
not modelled
100.0
17
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from oenococcus oeni
24 c1vftA_
not modelled
100.0
16
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of l-cycloserine-bound form of alanine2 racemase from d-cycloserine-producing streptomyces3 lavendulae
25 c3hurA_
not modelled
100.0
13
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from oenococcus oeni
26 c4lusD_
not modelled
100.0
13
PDB header: isomeraseChain: D: PDB Molecule: alanine racemase;PDBTitle: alanine racemase [clostridium difficile 630]
27 c4dzaA_
not modelled
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: lysine racemase;PDBTitle: crystal structure of a lysine racemase within internal aldimine2 linkage
28 d1rcqa2
not modelled
100.0
17
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
29 c3kw3B_
not modelled
100.0
13
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: crystal structure of alanine racemase from bartonella henselae with2 covalently bound pyridoxal phosphate
30 c2vd9A_
not modelled
100.0
15
PDB header: isomeraseChain: A: PDB Molecule: alanine racemase;PDBTitle: the crystal structure of alanine racemase from bacillus2 anthracis (ba0252) with bound l-ala-p
31 c6a2fB_
not modelled
100.0
18
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase, biosynthetic;PDBTitle: crystal structure of biosynthetic alanine racemase from pseudomonas2 aeruginosa
32 c4kbxA_
not modelled
100.0
22
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yhfx;PDBTitle: crystal structure of the pyridoxal-5'-phosphate dependent protein yhfx2 from escherichia coli
33 c4tloB_
not modelled
100.0
12
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: alanine racemase from acinetobacter baumannii
34 c2odoC_
not modelled
100.0
18
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of pseudomonas fluorescens alanine racemase
35 c2rjgC_
not modelled
100.0
15
PDB header: isomeraseChain: C: PDB Molecule: alanine racemase;PDBTitle: crystal structure of biosynthetic alaine racemase from escherichia2 coli
36 c3wqgB_
not modelled
100.0
19
PDB header: lyaseChain: B: PDB Molecule: d-threo-3-hydroxyaspartate dehydratase;PDBTitle: d-threo-3-hydroxyaspartate dehydratase c353a mutant in the metal-free2 form
37 c3anuA_
not modelled
100.0
14
PDB header: lyaseChain: A: PDB Molecule: d-serine dehydratase;PDBTitle: crystal structure of d-serine dehydratase from chicken kidney
38 c4v15B_
not modelled
100.0
21
PDB header: lyaseChain: B: PDB Molecule: d-threonine aldolase;PDBTitle: crystal structure of d-threonine aldolase from alcaligenes2 xylosoxidans
39 c3llxA_
not modelled
100.0
14
PDB header: isomeraseChain: A: PDB Molecule: predicted amino acid aldolase or racemase;PDBTitle: crystal structure of an ala racemase-like protein (il1761) from2 idiomarina loihiensis at 1.50 a resolution
40 c3gwqB_
not modelled
100.0
15
PDB header: lyaseChain: B: PDB Molecule: d-serine deaminase;PDBTitle: crystal structure of a putative d-serine deaminase (bxe_a4060) from2 burkholderia xenovorans lb400 at 2.00 a resolution
41 c4bhyB_
not modelled
99.9
17
PDB header: isomeraseChain: B: PDB Molecule: alanine racemase;PDBTitle: structure of alanine racemase from aeromonas hydrophila
42 c2j66A_
not modelled
99.9
12
PDB header: lyaseChain: A: PDB Molecule: btrk;PDBTitle: structural characterisation of btrk decarboxylase from2 butirosin biosynthesis
43 c2qghA_
not modelled
99.9
12
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase from helicobacter2 pylori complexed with l-lysine
44 c2p3eA_
not modelled
99.9
16
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of aq1208 from aquifex aeolicus
45 c3n2bD_
not modelled
99.9
18
PDB header: lyaseChain: D: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: 1.8 angstrom resolution crystal structure of diaminopimelate2 decarboxylase (lysa) from vibrio cholerae.
46 c4xg1C_
not modelled
99.9
16
PDB header: lyaseChain: C: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: psychromonas ingrahamii diaminopimelate decarboxylase with llp
47 c1njjC_
not modelled
99.8
8
PDB header: lyaseChain: C: PDB Molecule: ornithine decarboxylase;PDBTitle: crystal structure determination of t. brucei ornithine decarboxylase2 bound to d-ornithine and to g418
48 c3vabA_
not modelled
99.8
17
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase 1;PDBTitle: crystal structure of diaminopimelate decarboxylase from brucella2 melitensis bound to plp
49 c5x7nA_
not modelled
99.8
22
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal strcuture of meso-diaminopimelate decarboxylase (dapdc) from2 corynebacterium glutamicum
50 c6n2fB_
not modelled
99.8
18
PDB header: lyaseChain: B: PDB Molecule: diaminopimelate decarboxylase 2, chloroplastic;PDBTitle: meso-diaminopimelate decarboxylase from arabidopsis thaliana (isoform2 2)
51 c1tufA_
not modelled
99.8
16
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase from m.2 jannaschi
52 c5gjmB_
not modelled
99.8
17
PDB header: lyaseChain: B: PDB Molecule: lysine/ornithine decarboxylase;PDBTitle: crystal strcuture of lysine decarboxylase from selenomonas ruminantium2 in c2 space group
53 d1twia2
not modelled
99.8
17
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
54 c5bwaA_
not modelled
99.8
14
PDB header: lyase/lyase inhibitorChain: A: PDB Molecule: ornithine decarboxylase;PDBTitle: crystal structure of odc-plp-az1 ternary complex
55 d1hkva2
not modelled
99.8
22
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
56 c2nvaH_
not modelled
99.7
7
PDB header: lyaseChain: H: PDB Molecule: arginine decarboxylase, a207r protein;PDBTitle: the x-ray crystal structure of the paramecium bursaria chlorella virus2 arginine decarboxylase bound to agmatine
57 d7odca2
not modelled
99.7
14
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
58 c2pljA_
not modelled
99.7
15
PDB header: lyaseChain: A: PDB Molecule: lysine/ornithine decarboxylase;PDBTitle: crystal structure of lysine/ornithine decarboxylase complexed with2 putrescine from vibrio vulnificus
59 d1f3ta2
not modelled
99.7
10
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
60 c2o0tB_
not modelled
99.6
22
PDB header: lyaseChain: B: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: the three dimensional structure of diaminopimelate decarboxylase from2 mycobacterium tuberculosis reveals a tetrameric enzyme organisation
61 c1knwA_
not modelled
99.6
18
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure of diaminopimelate decarboxylase
62 c2yxxA_
not modelled
99.6
15
PDB header: lyaseChain: A: PDB Molecule: diaminopimelate decarboxylase;PDBTitle: crystal structure analysis of diaminopimelate decarboxylate (lysa)
63 d1d7ka2
not modelled
99.5
13
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
64 c3btnA_
not modelled
99.3
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: antizyme inhibitor 1;PDBTitle: crystal structure of antizyme inhibitor, an ornithine2 decarboxylase homologous protein
65 c2on3A_
not modelled
99.3
12
PDB header: lyaseChain: A: PDB Molecule: ornithine decarboxylase;PDBTitle: a structural insight into the inhibition of human and leishmania2 donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane
66 c3nzqB_
not modelled
99.2
17
PDB header: lyaseChain: B: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
67 c3n29A_
not modelled
99.2
12
PDB header: lyaseChain: A: PDB Molecule: carboxynorspermidine decarboxylase;PDBTitle: crystal structure of carboxynorspermidine decarboxylase complexed with2 norspermidine from campylobacter jejuni
68 d1knwa2
not modelled
99.1
18
Fold: TIM beta/alpha-barrelSuperfamily: PLP-binding barrelFamily: Alanine racemase-like, N-terminal domain
69 c1d7kB_
not modelled
99.1
13
PDB header: lyaseChain: B: PDB Molecule: human ornithine decarboxylase;PDBTitle: crystal structure of human ornithine decarboxylase at 2.1 angstroms2 resolution
70 c3n2oA_
not modelled
99.1
14
PDB header: lyaseChain: A: PDB Molecule: biosynthetic arginine decarboxylase;PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
71 c4aibC_
not modelled
99.0
15
PDB header: lyaseChain: C: PDB Molecule: ornithine decarboxylase;PDBTitle: crystal structure of ornithine decarboxylase from entamoeba2 histolytica.
72 c3nzpA_
not modelled
98.9
13
PDB header: lyaseChain: A: PDB Molecule: arginine decarboxylase;PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
73 c3mt1B_
not modelled
98.5
14
PDB header: lyaseChain: B: PDB Molecule: putative carboxynorspermidine decarboxylase protein;PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
74 c4nu7C_
not modelled
94.0
15
PDB header: isomeraseChain: C: PDB Molecule: ribulose-phosphate 3-epimerase;PDBTitle: 2.05 angstrom crystal structure of ribulose-phosphate 3-epimerase from2 toxoplasma gondii.
75 c1izcA_
not modelled
92.6
11
PDB header: lyaseChain: A: PDB Molecule: macrophomate synthase intermolecular diels-alderase;PDBTitle: crystal structure analysis of macrophomate synthase
76 d1izca_
not modelled
92.6
11
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase
77 d1tqxa_
not modelled
92.4
11
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
78 c3qz6A_
not modelled
89.9
13
PDB header: lyaseChain: A: PDB Molecule: hpch/hpai aldolase;PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
79 d1h1ya_
not modelled
89.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
80 d1dxea_
not modelled
88.5
16
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: HpcH/HpaI aldolase
81 c2vwtA_
not modelled
85.9
23
PDB header: lyaseChain: A: PDB Molecule: yfau, 2-keto-3-deoxy sugar aldolase;PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
82 c3ct7E_
not modelled
85.5
9
PDB header: isomeraseChain: E: PDB Molecule: d-allulose-6-phosphate 3-epimerase;PDBTitle: crystal structure of d-allulose 6-phosphate 3-epimerase2 from escherichia coli k-12
83 c3q94B_
not modelled
85.4
18
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase, class ii;PDBTitle: the crystal structure of fructose 1,6-bisphosphate aldolase from2 bacillus anthracis str. 'ames ancestor'
84 d1tqja_
not modelled
84.6
14
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
85 c4b5sB_
not modelled
84.3
19
PDB header: lyaseChain: B: PDB Molecule: 4-hydroxy-2-oxo-heptane-1,7-dioate aldolase;PDBTitle: crystal structures of divalent metal dependent pyruvate aldolase,2 hpai, in complex with pyruvate
86 c2v5jB_
not modelled
83.5
20
PDB header: lyaseChain: B: PDB Molecule: 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase;PDBTitle: apo class ii aldolase hpch
87 c6oviA_
not modelled
80.2
11
PDB header: lyaseChain: A: PDB Molecule: keto-deoxy-phosphogluconate aldolase;PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
88 c4mf4F_
not modelled
80.2
21
PDB header: lyaseChain: F: PDB Molecule: hpch/hpai aldolase/citrate lyase family protein;PDBTitle: crystal structure of a hpch/hpal aldolase/citrate lyase family protein2 from burkholderia cenocepacia j2315
89 c6r62A_
not modelled
73.5
17
PDB header: lyaseChain: A: PDB Molecule: hpch/hpai aldolase;PDBTitle: crystal structure of a class ii pyruvate aldolase from sphingomonas2 wittichii rw1 in complex with hydroxypyruvate
90 d1o4ua1
not modelled
73.4
17
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like
91 d2flia1
not modelled
70.8
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
92 d1gvfa_
not modelled
69.4
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase
93 d1rpxa_
not modelled
66.2
12
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: D-ribulose-5-phosphate 3-epimerase
94 c3inpA_
not modelled
63.3
14
PDB header: isomeraseChain: A: PDB Molecule: d-ribulose-phosphate 3-epimerase;PDBTitle: 2.05 angstrom resolution crystal structure of d-ribulose-phosphate 3-2 epimerase from francisella tularensis.
95 d1qpoa1
not modelled
54.6
26
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like
96 c3cu2A_
not modelled
51.3
14
PDB header: isomeraseChain: A: PDB Molecule: ribulose-5-phosphate 3-epimerase;PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
97 c2y85D_
not modelled
51.0
20
PDB header: isomeraseChain: D: PDB Molecule: phosphoribosyl isomerase a;PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
98 c2vkzH_
not modelled
50.8
10
PDB header: transferaseChain: H: PDB Molecule: fatty acid synthase subunit beta;PDBTitle: structure of the cerulenin-inhibited fungal fatty acid synthase type i2 multienzyme complex
99 c1yoeA_
not modelled
50.3
11
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein ybek;PDBTitle: crystal structure of a the e. coli pyrimidine nucleoside hydrolase2 ybek with bound ribose
100 d1y0ea_
not modelled
48.9
8
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: NanE-like
101 c1yadD_
not modelled
40.5
5
PDB header: transcriptionChain: D: PDB Molecule: regulatory protein teni;PDBTitle: structure of teni from bacillus subtilis
102 c4tv6A_
not modelled
39.9
16
PDB header: lyaseChain: A: PDB Molecule: 2-dehydro-3-deoxyglucarate aldolase;PDBTitle: crystal structure of citrate synthase variant sbng e151q
103 c2c3zA_
not modelled
39.7
14
PDB header: lyaseChain: A: PDB Molecule: indole-3-glycerol phosphate synthase;PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
104 c5mj7B_
not modelled
39.7
13
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: structure of the c. elegans nucleoside hydrolase
105 c3qc3B_
not modelled
38.7
12
PDB header: isomeraseChain: B: PDB Molecule: d-ribulose-5-phosphate-3-epimerase;PDBTitle: crystal structure of a d-ribulose-5-phosphate-3-epimerase (np_954699)2 from homo sapiens at 2.20 a resolution
106 d1kica_
not modelled
34.5
9
Fold: Nucleoside hydrolaseSuperfamily: Nucleoside hydrolaseFamily: Nucleoside hydrolase
107 c2w5fB_
not modelled
33.8
13
PDB header: hydrolaseChain: B: PDB Molecule: endo-1,4-beta-xylanase y;PDBTitle: high resolution crystallographic structure of the clostridium2 thermocellum n-terminal endo-1,4-beta-d-xylanase 10b (xyn10b) cbm22-3 1- gh10 modules complexed with xylohexaose
108 d1rvga_
not modelled
31.1
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class II FBP aldolase
109 c3ve9B_
not modelled
30.0
23
PDB header: lyaseChain: B: PDB Molecule: orotidine-5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5'-monophosphate decarboxylase from2 metallosphaera sedula
110 c3ewbX_
not modelled
27.8
10
PDB header: transferaseChain: X: PDB Molecule: 2-isopropylmalate synthase;PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
111 c1qpoA_
not modelled
23.8
27
PDB header: transferaseChain: A: PDB Molecule: quinolinate acid phosphoribosyl transferase;PDBTitle: quinolinate phosphoribosyl transferase (qaprtase) apo-enzyme from2 mycobacterium tuberculosis
112 c2b7pA_
not modelled
22.2
14
PDB header: transferaseChain: A: PDB Molecule: probable nicotinate-nucleotide pyrophosphorylase;PDBTitle: crystal structure of quinolinic acid phosphoribosyltransferase from2 helicobacter pylori with phthalic acid
113 c5u4nA_
not modelled
22.2
17
PDB header: lyaseChain: A: PDB Molecule: fructose-1;PDBTitle: crystal structure of a fructose-bisphosphate aldolase from neisseria2 gonorrhoeae
114 d1pkla2
not modelled
22.1
20
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Pyruvate kinase