| 1 | c1w59B_
|
|
 |
100.0 |
45 |
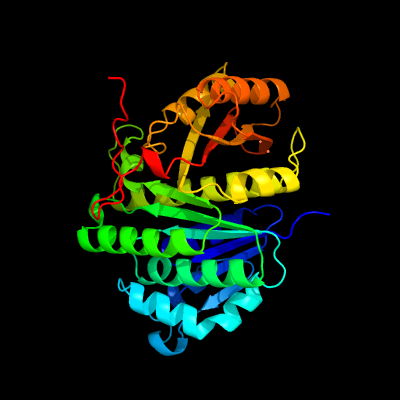
PDB header:cell division
Chain: B: PDB Molecule:cell division protein ftsz homolog 1;
PDBTitle: ftsz dimer, empty (m. jannaschii)
|
|
|
|
| 2 | c1w5fA_
|
|
 |
100.0 |
46 |
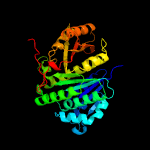
PDB header:cell division
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz, t7 mutated, domain swapped (t. maritima)
|
|
|
|
| 3 | c2r6r1_
|
|
 |
100.0 |
44 |
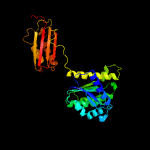
PDB header:cell cycle
Chain: 1: PDB Molecule:cell division protein ftsz;
PDBTitle: aquifex aeolicus ftsz
|
|
|
|
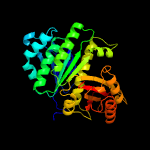
| 4 | c2vawA_
|
|
 |
100.0 |
52 |
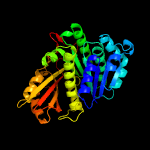
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: ftsz pseudomonas aeruginosa gdp
|
|
|
|
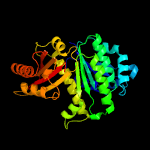
| 5 | c1ofuB_
|
|
 |
100.0 |
52 |
PDB header:bacterial cell division inhibitor
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of sula:ftsz from pseudomonas aeruginosa
|
|
|
|
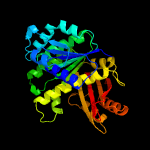
| 6 | c2vxyA_
|
|
 |
100.0 |
66 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: the structure of ftsz from bacillus subtilis at 1.7a2 resolution
|
|
|
|
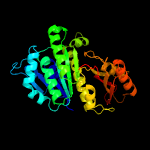
| 7 | c4dxdA_
|
|
 |
100.0 |
63 |
PDB header:cell cycle/inhibitor
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: staphylococcal aureus ftsz in complex with 723
|
|
|
|
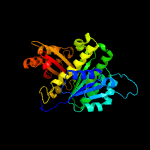
| 8 | c4e6eA_
|
|
 |
100.0 |
80 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of a putative cell division protein ftsz (tfu_1113)2 from thermobifida fusca yx-er1 at 2.22 a resolution (psi community3 target, van wezel g.p.)
|
|
|
|
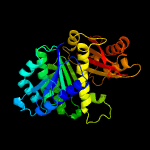
| 9 | c2rhoB_
|
|
 |
100.0 |
66 |
PDB header:cell cycle
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: synthetic gene encoded bacillus subtilis ftsz ncs dimer with bound gdp2 and gtp-gamma-s
|
|
|
|
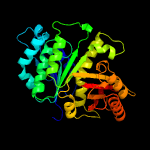
| 10 | c2q1yB_
|
|
 |
100.0 |
100 |
PDB header:cell cycle, signaling protein
Chain: B: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of cell division protein ftsz from mycobacterium2 tuberculosis in complex with gtp-gamma-s
|
|
|
|
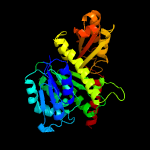
| 11 | c4b45A_
|
|
 |
100.0 |
23 |
PDB header:structural protein
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: cetz2 from haloferax volcanii - gtpgs bound protofilament
|
|
|
|
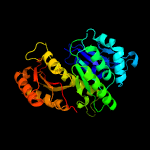
| 12 | c4b46A_
|
|
 |
100.0 |
25 |
PDB header:structural protein
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: cetz1 from haloferax volcanii - gdp bound monomer
|
|
|
|
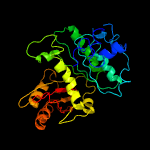
| 13 | c5v68A_
|
|
 |
100.0 |
97 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsz;
PDBTitle: crystal structure of cell division protein ftsz from mycobacterium2 tuberculosis bounded via the t9 loop
|
|
|
|
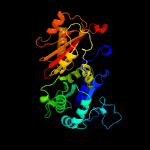
| 14 | c4ei8A_
|
|
 |
100.0 |
17 |
PDB header:replication
Chain: A: PDB Molecule:plasmid replication protein repx;
PDBTitle: crystal structure of bacillus cereus tubz, apo-form
|
|
|
|
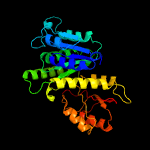
| 15 | c3v3tA_
|
|
 |
100.0 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:cell division gtpase ftsz, diverged;
PDBTitle: crystal structure of clostridium botulinum phage c-st tubz
|
|
|
|
| 16 | c3zidB_
|
|
 |
100.0 |
23 |
PDB header:gtp-binding protein
Chain: B: PDB Molecule:tubulin/ftsz, gtpase;
PDBTitle: cetz from methanosaeta thermophila strain dsm 6194
|
|
|
|
| 17 | d2vapa1
|
|
 |
100.0 |
50 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
|
| 18 | c3rb8A_
|
|
 |
100.0 |
13 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of the phage tubulin phuz(semet)-gdp
|
|
|
|
| 19 | d1ofua1
|
|
 |
100.0 |
57 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
|
| 20 | c3m8kA_
|
|
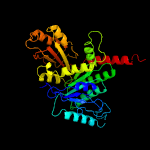 |
100.0 |
17 |
PDB header:structural protein
Chain: A: PDB Molecule:ftsz/tubulin-related protein;
PDBTitle: protein structure of type iii plasmid segregation tubz
|
|
|
|
| 21 | d1rq2a1 |
|
not modelled |
100.0 |
99 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 22 | d1w5fa1 |
|
not modelled |
100.0 |
53 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 23 | c3zbqA_ |
|
not modelled |
100.0 |
16 |
PDB header:viral protein
Chain: A: PDB Molecule:phikz039;
PDBTitle: protofilament of tubz from bacteriophage phikz
|
|
|
| 24 | d1w5fa2 |
|
not modelled |
100.0 |
35 |
Fold:Bacillus chorismate mutase-like
Superfamily:Tubulin C-terminal domain-like
Family:Tubulin, C-terminal domain |
|
|
| 25 | d2vapa2 |
|
not modelled |
100.0 |
39 |
Fold:Bacillus chorismate mutase-like
Superfamily:Tubulin C-terminal domain-like
Family:Tubulin, C-terminal domain |
|
|
| 26 | d1rq2a2 |
|
not modelled |
100.0 |
100 |
Fold:Bacillus chorismate mutase-like
Superfamily:Tubulin C-terminal domain-like
Family:Tubulin, C-terminal domain |
|
|
| 27 | d1ofua2 |
|
not modelled |
100.0 |
44 |
Fold:Bacillus chorismate mutase-like
Superfamily:Tubulin C-terminal domain-like
Family:Tubulin, C-terminal domain |
|
|
| 28 | c5flzC_ |
|
not modelled |
99.8 |
13 |
PDB header:cell cycle
Chain: C: PDB Molecule:tubulin gamma chain;
PDBTitle: cryo-em structure of gamma-tusc oligomers in a closed conformation
|
|
|
| 29 | c2p4nB_ |
|
not modelled |
99.7 |
15 |
PDB header:transport protein
Chain: B: PDB Molecule:tubulin beta chain;
PDBTitle: human monomeric kinesin (1bg2) and bovine tubulin (1jff) docked into2 the 9-angstrom cryo-em map of nucleotide-free kinesin complexed to3 the microtubule
|
|
|
| 30 | c3edlA_ |
|
not modelled |
99.7 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:alpha-tubulin;
PDBTitle: kinesin13-microtubule ring complex
|
|
|
| 31 | c1z5wA_ |
|
not modelled |
99.7 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:tubulin gamma-1 chain;
PDBTitle: crystal structure of gamma-tubulin bound to gtp
|
|
|
| 32 | c2btqB_ |
|
not modelled |
99.7 |
17 |
PDB header:structural protein
Chain: B: PDB Molecule:tubulin btubb;
PDBTitle: structure of btubab heterodimer from prosthecobacter dejongeii
|
|
|
| 33 | c2btoA_ |
|
not modelled |
99.7 |
19 |
PDB header:cytoskeletal protein
Chain: A: PDB Molecule:tubulin btuba;
PDBTitle: structure of btuba from prosthecobacter dejongeii
|
|
|
| 34 | d1tubb1 |
|
not modelled |
99.6 |
19 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 35 | d2btoa1 |
|
not modelled |
99.6 |
18 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 36 | d1tuba1 |
|
not modelled |
99.6 |
16 |
Fold:Tubulin nucleotide-binding domain-like
Superfamily:Tubulin nucleotide-binding domain-like
Family:Tubulin, GTPase domain |
|
|
| 37 | d1i10a1 |
|
not modelled |
97.1 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 38 | c2v65A_ |
|
not modelled |
96.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase a chain;
PDBTitle: apo ldh from the psychrophile c. gunnari
|
|
|
| 39 | d1llda1 |
|
not modelled |
96.5 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 40 | c1pzfD_ |
|
not modelled |
96.5 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:lactate dehydrogenase;
PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate
|
|
|
| 41 | c4bgvB_ |
|
not modelled |
96.4 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: 1.8 a resolution structure of the malate dehydrogenase from2 picrophilus torridus in its apo form
|
|
|
| 42 | c1ldbA_ |
|
not modelled |
96.4 |
26 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-l-lactate dehydrogenase;
PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
|
|
|
| 43 | d1pzga1 |
|
not modelled |
96.4 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 44 | c1ez4B_ |
|
not modelled |
96.3 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of non-allosteric l-lactate dehydrogenase2 from lactobacillus pentosus at 2.3 angstrom resolution
|
|
|
| 45 | d9ldta1 |
|
not modelled |
96.2 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 46 | d1ldna1 |
|
not modelled |
96.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 47 | c3d0oA_ |
|
not modelled |
96.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of lactate dehydrogenase from staphylococcus aureus
|
|
|
| 48 | d1u8xx1 |
|
not modelled |
96.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 49 | c1u4sA_ |
|
not modelled |
96.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
|
|
|
| 50 | c3pqeD_ |
|
not modelled |
96.0 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus subtilis2 with h171c mutation
|
|
|
| 51 | c3wsvC_ |
|
not modelled |
96.0 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of minor l-lactate dehydrogenase from enterococcus2 mundtii in the ligands-unbound form
|
|
|
| 52 | c6or9B_ |
|
not modelled |
95.9 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: structure of l-lactate dehydrogenase from trichoplusia ni
|
|
|
| 53 | d1ldma1 |
|
not modelled |
95.9 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 54 | d2cmda1 |
|
not modelled |
95.7 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 55 | c8ldhA_ |
|
not modelled |
95.7 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate dehydrogenase
|
|
|
| 56 | c1a5zA_ |
|
not modelled |
95.6 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: lactate dehydrogenase from thermotoga maritima (tmldh)
|
|
|
| 57 | d1y6ja1 |
|
not modelled |
95.6 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 58 | d1i0za1 |
|
not modelled |
95.6 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 59 | c3fefB_ |
|
not modelled |
95.6 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative glucosidase lpld, alpha-galacturonidase;
PDBTitle: crystal structure of putative glucosidase lpld from bacillus subtilis
|
|
|
| 60 | c4ln1B_ |
|
not modelled |
95.6 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase 1;
PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus cereus atcc2 14579 complexed with calcium, nysgrc target 029452
|
|
|
| 61 | d2ldxa1 |
|
not modelled |
95.5 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 62 | c1obbB_ |
|
not modelled |
95.5 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucosidase;
PDBTitle: alpha-glucosidase a, agla, from thermotoga maritima in complex with2 maltose and nad+
|
|
|
| 63 | c6ct6B_ |
|
not modelled |
95.5 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from eimeria maxima with2 nadh and oxamate
|
|
|
| 64 | c3dl2A_ |
|
not modelled |
95.5 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ubiquitin-conjugating enzyme e2 variant 3;
PDBTitle: hexagonal structure of the ldh domain of human ubiquitin-2 conjugating enzyme e2-like isoform a
|
|
|
| 65 | d1ez4a1 |
|
not modelled |
95.4 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 66 | c1lldA_ |
|
not modelled |
95.4 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: molecular basis of allosteric activation of bacterial l-lactate2 dehydrogenase
|
|
|
| 67 | d1hyha1 |
|
not modelled |
95.3 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 68 | d1mlda1 |
|
not modelled |
95.3 |
20 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 69 | c3gucB_ |
|
not modelled |
95.3 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:ubiquitin-like modifier-activating enzyme 5;
PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
|
|
|
| 70 | c1hyhA_ |
|
not modelled |
95.3 |
25 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
|
|
| 71 | c3h9gA_ |
|
not modelled |
95.1 |
20 |
PDB header:transferase/antibiotic
Chain: A: PDB Molecule:mccb protein;
PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
|
|
|
| 72 | c3p7mC_ |
|
not modelled |
95.1 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of putative lactate dehydrogenase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 73 | d1a5za1 |
|
not modelled |
95.1 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 74 | c4q3nA_ |
|
not modelled |
95.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:mgs-m5;
PDBTitle: crystal structure of mgs-m5, a lactate dehydrogenase enzyme from a2 medee basin deep-sea metagenome library
|
|
|
| 75 | c2v6bB_ |
|
not modelled |
95.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of lactate dehydrogenase from deinococcus2 radiodurans (apo form)
|
|
|
| 76 | c1y6jA_ |
|
not modelled |
95.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: l-lactate dehydrogenase from clostridium thermocellum cth-1135
|
|
|
| 77 | d1up7a1 |
|
not modelled |
95.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 78 | c1hygA_ |
|
not modelled |
94.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate/malate dehydrogenase;
PDBTitle: crystal structure of mj0490 gene product, the family of2 lactate/malate dehydrogenase
|
|
|
| 79 | c1ojuA_ |
|
not modelled |
94.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: 2.8 a resolution structure of malate dehydrogenase from2 archaeoglobus fulgidus in complex with etheno-nad.
|
|
|
| 80 | d7mdha1 |
|
not modelled |
94.8 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 81 | c3ic5A_ |
|
not modelled |
94.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative saccharopine dehydrogenase;
PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
|
|
|
| 82 | c2e37B_ |
|
not modelled |
94.7 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: structure of tt0471 protein from thermus thermophilus
|
|
|
| 83 | c2dfdD_ |
|
not modelled |
94.7 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of human malate dehydrogenase type 2
|
|
|
| 84 | d5ldha1 |
|
not modelled |
94.7 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 85 | d1obba1 |
|
not modelled |
94.7 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 86 | c2hjrK_ |
|
not modelled |
94.6 |
26 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
|
|
|
| 87 | c4plcA_ |
|
not modelled |
94.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of ancestral apicomplexan lactate dehydrogenase with2 malate.
|
|
|
| 88 | d1jw9b_ |
|
not modelled |
94.2 |
22 |
Fold:Activating enzymes of the ubiquitin-like proteins
Superfamily:Activating enzymes of the ubiquitin-like proteins
Family:Molybdenum cofactor biosynthesis protein MoeB |
|
|
| 89 | d1p3da1 |
|
not modelled |
94.2 |
24 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 90 | d1ojua1 |
|
not modelled |
94.1 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 91 | c1u8xX_ |
|
not modelled |
94.0 |
22 |
PDB header:hydrolase
Chain: X: PDB Molecule:maltose-6'-phosphate glucosidase;
PDBTitle: crystal structure of glva from bacillus subtilis, a metal-requiring,2 nad-dependent 6-phospho-alpha-glucosidase
|
|
|
| 92 | c2ldxA_ |
|
not modelled |
93.9 |
24 |
PDB header:oxidoreductase(choh(d)-nad(a))
Chain: A: PDB Molecule:apo-lactate dehydrogenase;
PDBTitle: characterization of the antigenic sites on the refined 3-2 angstroms resolution structure of mouse testicular lactate3 dehydrogenase c4
|
|
|
| 93 | c1gv1D_ |
|
not modelled |
93.8 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for thermophilic protein stability:2 structures of thermophilic and mesophilic malate3 dehydrogenases
|
|
|
| 94 | d1gv0a1 |
|
not modelled |
93.7 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 95 | c1mldA_ |
|
not modelled |
93.6 |
19 |
PDB header:oxidoreductase(nad(a)-choh(d))
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
|
|
|
| 96 | d1t2da1 |
|
not modelled |
93.6 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 97 | d1uxja1 |
|
not modelled |
93.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 98 | d1pjqa1 |
|
not modelled |
93.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Siroheme synthase N-terminal domain-like |
|
|
| 99 | c3gviB_ |
|
not modelled |
93.5 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of lactate/malate dehydrogenase from2 brucella melitensis in complex with adp
|
|
|
| 100 | d1s6ya1 |
|
not modelled |
93.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 101 | c3triB_ |
|
not modelled |
93.4 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:pyrroline-5-carboxylate reductase;
PDBTitle: structure of a pyrroline-5-carboxylate reductase (proc) from coxiella2 burnetii
|
|
|
| 102 | c1ur5C_ |
|
not modelled |
93.4 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:malate dehydrogenase;
PDBTitle: stabilization of a tetrameric malate dehydrogenase by introduction of2 a disulfide bridge at the dimer/dimer interface
|
|
|
| 103 | c6qssA_ |
|
not modelled |
93.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of ignicoccus islandicus malate dehydrogenase co-2 crystallized with 10 mm tb-xo4
|
|
|
| 104 | d1guza1 |
|
not modelled |
93.3 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 105 | c4uupB_ |
|
not modelled |
93.3 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: reconstructed ancestral trichomonad malate dehydrogenase in2 complex with nadh, so4, and po4
|
|
|
| 106 | c6ihdA_ |
|
not modelled |
93.2 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal strcuture of malate dehydrogenase from metallosphaera sedula
|
|
|
| 107 | c1smkD_ |
|
not modelled |
93.0 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:malate dehydrogenase, glyoxysomal;
PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
|
|
|
| 108 | c1sevA_ |
|
not modelled |
93.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase, glyoxysomal precursor;
PDBTitle: mature and translocatable forms of glyoxysomal malate2 dehydrogenase have different activities and stabilities3 but similar crystal structures
|
|
|
| 109 | c1s6yA_ |
|
not modelled |
93.0 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: 2.3a crystal structure of phospho-beta-glucosidase
|
|
|
| 110 | d1llca1 |
|
not modelled |
93.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 111 | c5nfrI_ |
|
not modelled |
92.9 |
23 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from plasmodium falciparum2 (pfmdh)
|
|
|
| 112 | c6pblA_ |
|
not modelled |
92.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from legionella pneumophila2 philadelphia 1
|
|
|
| 113 | c2axqA_ |
|
not modelled |
92.8 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:saccharopine dehydrogenase;
PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
|
|
|
| 114 | c2fnzA_ |
|
not modelled |
92.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lactate dehydrogenase;
PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
|
|
|
| 115 | c1up6F_ |
|
not modelled |
92.7 |
24 |
PDB header:hydrolase
Chain: F: PDB Molecule:6-phospho-beta-glucosidase;
PDBTitle: structure of the 6-phospho-beta glucosidase from thermotoga2 maritima at 2.55 angstrom resolution in the tetragonal form3 with manganese, nad+ and glucose-6-phosphate
|
|
|
| 116 | c5l78A_ |
|
not modelled |
92.6 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-aminoadipic semialdehyde synthase, mitochondrial;
PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
|
|
|
| 117 | d1y7ta1 |
|
not modelled |
92.5 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 118 | c4tvoB_ |
|
not modelled |
92.4 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of malate dehydrogenase from mycobacterium tuberculosis
|
|
|
| 119 | c2nvuB_ |
|
not modelled |
92.4 |
24 |
PDB header:protein turnover, ligase
Chain: B: PDB Molecule:maltose binding protein/nedd8-activating enzyme
PDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
|
|
|
| 120 | c1y8qD_ |
|
not modelled |
92.3 |
21 |
PDB header:ligase
Chain: D: PDB Molecule:ubiquitin-like 2 activating enzyme e1b;
PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
|
|
|