1 c2xjaD_
100.0
99
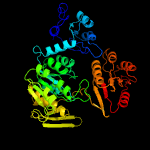
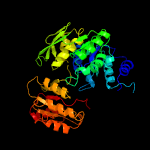
PDB header: ligaseChain: D: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--2,6-PDBTitle: structure of mure from m.tuberculosis with dipeptide and adp
2 c4c13A_
100.0
29
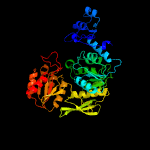
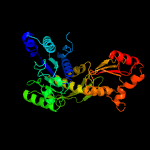
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--l-lysine ligase;PDBTitle: x-ray crystal structure of staphylococcus aureus mure with udp-murnac-2 ala-glu-lys
3 c1e8cB_
100.0
37
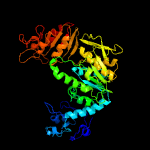
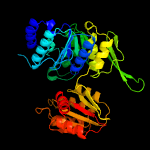
PDB header: ligaseChain: B: PDB Molecule: udp-n-acetylmuramoylalanyl-d-glutamate--2,6-diaminopimelatePDBTitle: structure of mure the udp-n-acetylmuramyl tripeptide synthetase from2 e. coli
4 c2wtzC_
100.0
98
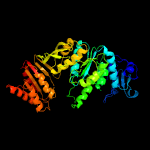
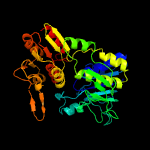
PDB header: ligaseChain: C: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate-PDBTitle: mure ligase of mycobacterium tuberculosis
5 c4bubA_
100.0
32
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--ld-lysinePDBTitle: crystal structure of mure ligase from thermotoga maritima2 in complex with adp
6 c4qdiA_
100.0
22
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-tripeptide--d-alanyl-d-alanine ligase;PDBTitle: crystal structure ii of murf from acinetobacter baumannii
7 c2am1A_
100.0
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine-d-glutamyl-lysine-d-alanyl-d-PDBTitle: sp protein ligand 1
8 c3zl8A_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-tripeptide--d-alanyl-d-alaninePDBTitle: crystal structure of murf ligase from thermotoga maritima2 in complex with adp
9 c4cvkA_
100.0
28
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-tripeptide--d-alanyl-d-alaninePDBTitle: pamurf in complex with udp-murnac-tripeptide (mdap)
10 c1gg4A_
100.0
28
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanyl-d-glutamyl-2,6-diaminopimelate-PDBTitle: crystal structure of escherichia coli udpmurnac-tripeptide d-alanyl-d-2 alanine-adding enzyme (murf) at 2.3 angstrom resolution
11 c3lk7A_
100.0
23
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine--d-glutamate ligase;PDBTitle: the crystal structure of udp-n-acetylmuramoylalanine-d-glutamate2 (murd) ligase from streptococcus agalactiae to 1.5a
12 c2vosA_
100.0
24
PDB header: ligaseChain: A: PDB Molecule: folylpolyglutamate synthase protein folc;PDBTitle: mycobacterium tuberculosis folylpolyglutamate synthase2 complexed with adp
13 c2f00A_
100.0
18
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
14 c3hn7A_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: crystal structure of a murein peptide ligase mpl (psyc_0032) from2 psychrobacter arcticus 273-4 at 1.65 a resolution
15 c1o5zA_
100.0
19
PDB header: ligaseChain: A: PDB Molecule: folylpolyglutamate synthase/dihydrofolate synthase;PDBTitle: crystal structure of folylpolyglutamate synthase (tm0166) from2 thermotoga maritima at 2.10 a resolution
16 c2gc6A_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: folylpolyglutamate synthase;PDBTitle: s73a mutant of l. casei fpgs
17 c1w78A_
100.0
23
PDB header: synthaseChain: A: PDB Molecule: folc bifunctional protein;PDBTitle: e.coli folc in complex with dhpp and adp
18 c3uagA_
100.0
25
PDB header: ligaseChain: A: PDB Molecule: protein (udp-n-acetylmuramoyl-l-alanine:d-PDBTitle: udp-n-acetylmuramoyl-l-alanine:d-glutamate ligase
19 c1gqqA_
100.0
17
PDB header: cell wall biosynthesisChain: A: PDB Molecule: udp-n-acetylmuramate-l-alanine ligase;PDBTitle: murc - crystal structure of the apo-enzyme from haemophilus influenzae
20 c1j6uA_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
21 c4bucA_
not modelled
100.0
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoylalanine--d-glutamate ligase;PDBTitle: crystal structure of murd ligase from thermotoga maritima in apo form
22 c3n2aA_
not modelled
100.0
23
PDB header: ligaseChain: A: PDB Molecule: bifunctional folylpolyglutamate synthase/dihydrofolatePDBTitle: crystal structure of bifunctional folylpolyglutamate2 synthase/dihydrofolate synthase from yersinia pestis co92
23 c6gs2B_
not modelled
100.0
16
PDB header: biosynthetic proteinChain: B: PDB Molecule: sa1708 protein;PDBTitle: crystal structure of the gatd/murt enzyme complex from staphylococcus2 aureus
24 c6fqbD_
not modelled
100.0
20
PDB header: ligaseChain: D: PDB Molecule: mur ligase family protein;PDBTitle: murt/gatd peptidoglycan amidotransferase complex from streptococcus2 pneumoniae r6
25 d1e8ca3
not modelled
100.0
36
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
26 c5vvwA_
not modelled
100.0
23
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: structure of murc from pseudomonas aeruginosa
27 d2gc6a2
not modelled
100.0
20
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: Folylpolyglutamate synthetase
28 d1p3da3
not modelled
100.0
20
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
29 c3eagA_
not modelled
100.0
19
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
30 c6cauA_
not modelled
100.0
22
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
31 d2jfga3
not modelled
100.0
26
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
32 d1o5za2
not modelled
100.0
16
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: Folylpolyglutamate synthetase
33 d1e8ca2
not modelled
100.0
45
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
34 d1j6ua3
not modelled
100.0
20
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
35 d1gg4a4
not modelled
100.0
22
Fold: Ribokinase-likeSuperfamily: MurD-like peptide ligases, catalytic domainFamily: MurCDEF
36 d1gg4a3
not modelled
99.9
32
Fold: MurF and HprK N-domain-likeSuperfamily: MurE/MurF N-terminal domainFamily: MurE/MurF N-terminal domain
37 d1e8ca1
not modelled
99.9
31
Fold: MurF and HprK N-domain-likeSuperfamily: MurE/MurF N-terminal domainFamily: MurE/MurF N-terminal domain
38 d1p3da2
not modelled
99.8
17
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
39 c3mvnA_
not modelled
99.8
27
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-medo-PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamyl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
40 d1j6ua2
not modelled
99.8
26
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
41 d2jfga2
not modelled
99.8
20
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
42 d1gg4a1
not modelled
99.7
26
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain
43 d1o5za1
not modelled
99.7
27
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: Folylpolyglutamate synthetase, C-terminal domain
44 d2gc6a1
not modelled
99.6
24
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: Folylpolyglutamate synthetase, C-terminal domain
45 c3pmoA_
not modelled
98.1
25
PDB header: transferaseChain: A: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosamine n-acyltransferase;PDBTitle: the structure of lpxd from pseudomonas aeruginosa at 1.3 a resolution
46 c3eh0C_
not modelled
97.7
23
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosamine n-PDBTitle: crystal structure of lpxd from escherichia coli
47 c2iu9C_
not modelled
97.1
12
PDB header: transferaseChain: C: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] glucosaminePDBTitle: chlamydia trachomatis lpxd with 100mm udpglcnac (complex ii)
48 c4e75A_
not modelled
97.1
16
PDB header: transferaseChain: A: PDB Molecule: udp-3-o-acylglucosamine n-acyltransferase;PDBTitle: structure of lpxd from acinetobacter baumannii at 2.85a resolution2 (p21 form)
49 d1a7ja_
not modelled
96.5
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase
50 c3fmfA_
not modelled
96.3
28
PDB header: ligaseChain: A: PDB Molecule: dethiobiotin synthetase;PDBTitle: crystal structure of mycobacterium tuberculosis dethiobiotin2 synthetase complexed with 7,8 diaminopelargonic acid carbamate
51 c2j37W_
not modelled
96.2
20
PDB header: ribosomeChain: W: PDB Molecule: signal recognition particle 54 kda protein (srp54);PDBTitle: model of mammalian srp bound to 80s rncs
52 c3b9qA_
not modelled
96.1
24
PDB header: protein transportChain: A: PDB Molecule: chloroplast srp receptor homolog, alpha subunitPDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
53 c2og2A_
not modelled
96.0
26
PDB header: protein transportChain: A: PDB Molecule: putative signal recognition particle receptor;PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
54 c2cnwF_
not modelled
95.9
27
PDB header: signal recognitionChain: F: PDB Molecule: cell division protein ftsy;PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
55 c5l3sF_
not modelled
95.8
19
PDB header: protein transportChain: F: PDB Molecule: signal recognition particle receptor ftsy;PDBTitle: structure of the gtpase heterodimer of crenarchaeal srp54 and ftsy
56 c6cy1B_
not modelled
95.8
27
PDB header: signaling proteinChain: B: PDB Molecule: signal recognition particle receptor ftsy;PDBTitle: crystal structure of signal recognition particle receptor ftsy from2 elizabethkingia anophelis
57 c3tqcB_
not modelled
95.6
27
PDB header: transferaseChain: B: PDB Molecule: pantothenate kinase;PDBTitle: structure of the pantothenate kinase (coaa) from coxiella burnetii
58 c3dm5A_
not modelled
95.6
22
PDB header: rna binding protein, transport proteinChain: A: PDB Molecule: signal recognition 54 kda protein;PDBTitle: structures of srp54 and srp19, the two proteins assembling the2 ribonucleic core of the signal recognition particle from the archaeon3 pyrococcus furiosus.
59 c3of5A_
not modelled
95.5
19
PDB header: ligaseChain: A: PDB Molecule: dethiobiotin synthetase;PDBTitle: crystal structure of a dethiobiotin synthetase from francisella2 tularensis subsp. tularensis schu s4
60 c2qy9A_
not modelled
95.5
22
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
61 c1qzwC_
not modelled
95.2
20
PDB header: signaling protein/rnaChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
62 c5l3qB_
not modelled
95.2
22
PDB header: protein transportChain: B: PDB Molecule: signal recognition particle receptor subunit alpha;PDBTitle: structure of the gtpase heterodimer of human srp54 and sralpha
63 c2f1rA_
not modelled
95.1
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesisPDBTitle: crystal structure of molybdopterin-guanine biosynthesis2 protein b (mobb)
64 c1vmaA_
not modelled
95.0
24
PDB header: protein transportChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
65 d1byia_
not modelled
95.0
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
66 d1xjca_
not modelled
94.8
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
67 c4dzzB_
not modelled
94.7
30
PDB header: unknown functionChain: B: PDB Molecule: plasmid partitioning protein parf;PDBTitle: structure of parf-adp, crystal form 1
68 c4nkrB_
not modelled
94.5
19
PDB header: unknown functionChain: B: PDB Molecule: molybdopterin-guanine dinucleotide biosynthesis protein b;PDBTitle: the crystal structure of bacillus subtilis mobb
69 c3cioA_
not modelled
94.4
20
PDB header: signaling protein, transferaseChain: A: PDB Molecule: tyrosine-protein kinase etk;PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
70 c2bekB_
not modelled
94.3
29
PDB header: chromosome segregationChain: B: PDB Molecule: segregation protein;PDBTitle: structure of the bacterial chromosome segregation protein soj
71 c2yhsA_
not modelled
94.2
20
PDB header: cell cycleChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of the e. coli srp receptor ftsy
72 d1vmaa2
not modelled
94.2
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
73 c2qmoA_
not modelled
94.2
34
PDB header: ligaseChain: A: PDB Molecule: dethiobiotin synthetase;PDBTitle: crystal structure of dethiobiotin synthetase (biod) from helicobacter2 pylori
74 c2iy3A_
not modelled
94.1
21
PDB header: rna-bindingChain: A: PDB Molecule: signal recognition particle protein,signal recognitionPDBTitle: structure of the e. coli signal regognition particle
75 d2qy9a2
not modelled
93.9
28
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
76 d2afhe1
not modelled
93.9
31
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
77 c2ozeA_
not modelled
93.9
16
PDB header: dna binding proteinChain: A: PDB Molecule: orf delta';PDBTitle: the crystal structure of delta protein of psm19035 from2 streptoccocus pyogenes
78 c1hyqA_
not modelled
93.7
27
PDB header: cell cycleChain: A: PDB Molecule: cell division inhibitor (mind-1);PDBTitle: mind bacterial cell division regulator from a. fulgidus
79 d1hyqa_
not modelled
93.7
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
80 c3la6P_
not modelled
93.7
18
PDB header: transferaseChain: P: PDB Molecule: tyrosine-protein kinase wzc;PDBTitle: octameric kinase domain of the e. coli tyrosine kinase wzc with bound2 adp
81 c2vedA_
not modelled
93.7
20
PDB header: transferaseChain: A: PDB Molecule: membrane protein capa1, protein tyrosine kinase;PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
82 d2g0ta1
not modelled
93.6
20
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
83 c4ru8C_
not modelled
93.6
50
PDB header: hydrolaseChain: C: PDB Molecule: uncharacterized protein;PDBTitle: structure of pnob8 para with amppnp
84 c5j1jA_
not modelled
93.6
37
PDB header: transcriptionChain: A: PDB Molecule: site-determining protein;PDBTitle: structure of flen-amppnp complex
85 d1sq5a_
not modelled
93.5
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase
86 d1rz3a_
not modelled
93.4
27
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase
87 c3c8uA_
not modelled
93.3
21
PDB header: transferaseChain: A: PDB Molecule: fructokinase;PDBTitle: crystal structure of putative fructose transport system kinase2 (yp_612366.1) from silicibacter sp. tm1040 at 1.95 a resolution
88 c2v3cC_
not modelled
93.1
34
PDB header: signaling proteinChain: C: PDB Molecule: signal recognition 54 kda protein;PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
89 d1iona_
not modelled
93.1
34
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
90 d1okkd2
not modelled
93.1
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
91 c2xj9B_
not modelled
93.1
26
PDB header: replicationChain: B: PDB Molecule: mipz;PDBTitle: dimer structure of the bacterial cell division regulator mipz
92 c4pfsA_
not modelled
93.0
37
PDB header: ligaseChain: A: PDB Molecule: cobyrinic acid a,c-diamide synthase;PDBTitle: crystal structure of cobyrinic acid a,c-diamide synthase from2 mycobacterium smegmatis
93 c4v02B_
not modelled
92.9
44
PDB header: cell cycleChain: B: PDB Molecule: site-determining protein;PDBTitle: minc:mind cell division protein complex, aquifex aeolicus
94 c6g2gA_
not modelled
92.9
28
PDB header: cytosolic proteinChain: A: PDB Molecule: cytosolic fe-s cluster assembly factor cfd1;PDBTitle: fe-s assembly cfd1
95 c2ph1A_
not modelled
92.8
28
PDB header: ligand binding proteinChain: A: PDB Molecule: nucleotide-binding protein;PDBTitle: crystal structure of nucleotide-binding protein af2382 from2 archaeoglobus fulgidus, northeast structural genomics target gr165
96 d1ihua2
not modelled
92.7
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
97 c4rz3B_
not modelled
92.6
24
PDB header: structural proteinChain: B: PDB Molecule: site-determining protein;PDBTitle: crystal structure of the mind-like atpase flhg
98 c3dmdA_
not modelled
92.6
25
PDB header: transport proteinChain: A: PDB Molecule: signal recognition particle receptor;PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
99 c1zu4A_
not modelled
92.5
13
PDB header: protein transportChain: A: PDB Molecule: ftsy;PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
100 d1np6a_
not modelled
91.9
32
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
101 c4xc8B_
not modelled
91.9
25
PDB header: isomeraseChain: B: PDB Molecule: isobutyryl-coa mutase fused;PDBTitle: isobutyryl-coa mutase fused with bound butyryl-coa, gdp, and mg and2 without cobalamin (apo-icmf/gdp)
102 c2q9cA_
not modelled
91.4
22
PDB header: signaling proteinChain: A: PDB Molecule: cell division protein ftsy;PDBTitle: structure of ftsy:gmppnp with mgcl complex
103 c3vx3A_
not modelled
91.4
25
PDB header: adp binding proteinChain: A: PDB Molecule: atpase involved in chromosome partitioning, para/mindPDBTitle: crystal structure of [nife] hydrogenase maturation protein hypb from2 thermococcus kodakarensis kod1
104 c3endA_
not modelled
91.4
30
PDB header: oxidoreductaseChain: A: PDB Molecule: light-independent protochlorophyllide reductasePDBTitle: crystal structure of the l protein of rhodobacter2 sphaeroides light-independent protochlorophyllide3 reductase (bchl) with mgadp bound: a homologue of the4 nitrogenase fe protein
105 d1g3qa_
not modelled
91.2
38
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
106 c4ak9A_
not modelled
91.0
25
PDB header: protein transportChain: A: PDB Molecule: cpftsy;PDBTitle: structure of chloroplast ftsy from physcomitrella patens
107 d1qzxa3
not modelled
90.6
30
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like
108 d1odfa_
not modelled
90.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Phosphoribulokinase/pantothenate kinase
109 c5zmfA_
not modelled
90.2
15
PDB header: hydrolase/transport proteinChain: A: PDB Molecule: atpase arsa1;PDBTitle: amppnp complex of c. reinhardtii arsa1
110 c2gesA_
not modelled
90.1
21
PDB header: transferaseChain: A: PDB Molecule: pantothenate kinase;PDBTitle: pantothenate kinase from mycobacterium tuberculosis (mtpank) in2 complex with a coenzyme a derivative, form-i (rt)
111 c3k9gA_
not modelled
90.0
27
PDB header: biosynthetic proteinChain: A: PDB Molecule: pf-32 protein;PDBTitle: crystal structure of a plasmid partition protein from borrelia2 burgdorferi at 2.25a resolution, iodide soak
112 c5l3rC_
not modelled
89.9
28
PDB header: protein transportChain: C: PDB Molecule: signal recognition particle 54 kda protein, chloroplastic;PDBTitle: structure of the gtpase heterodimer of chloroplast srp54 and ftsy from2 arabidopsis thaliana
113 c5gafi_
not modelled
89.5
21
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l10;PDBTitle: rnc in complex with srp
114 c3q9lB_
not modelled
89.4
14
PDB header: cell cycle, hydrolaseChain: B: PDB Molecule: septum site-determining protein mind;PDBTitle: the structure of the dimeric e.coli mind-atp complex
115 c3fkqA_
not modelled
88.9
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ntrc-like two-domain protein;PDBTitle: crystal structure of ntrc-like two-domain protein (rer070207001320)2 from eubacterium rectale at 2.10 a resolution
116 d2ioja1
not modelled
88.9
11
Fold: MurF and HprK N-domain-likeSuperfamily: HprK N-terminal domain-likeFamily: DRTGG domain
117 c6iucC_
not modelled
88.7
34
PDB header: dna binding protein/dnaChain: C: PDB Molecule: spooj regulator (soj);PDBTitle: structure of helicobacter pylori soj-atp complex bound to dna
118 c3ez6B_
not modelled
88.5
25
PDB header: dna binding proteinChain: B: PDB Molecule: plasmid partition protein a;PDBTitle: structure of para-adp complex:tetragonal form
119 d1nksa_
not modelled
88.4
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nucleotide and nucleoside kinases
120 c2obnA_
not modelled
88.1
16
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a duf1611 family protein (ava_3511) from anabaena2 variabilis atcc 29413 at 2.30 a resolution