| 1 | c2b81D_
|
|
 |
100.0 |
23 |
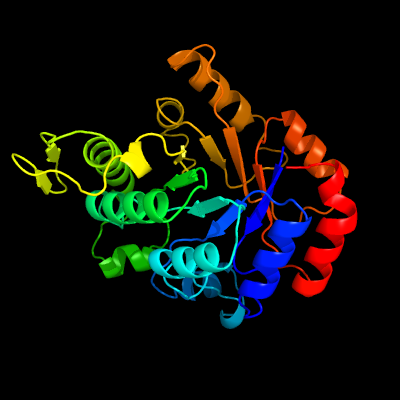
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
|
| 2 | c5tlcA_
|
|
 |
100.0 |
23 |
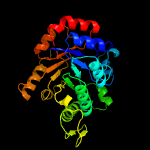
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
| 3 | d1lucb_
|
|
 |
100.0 |
16 |
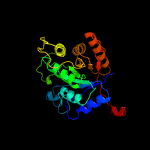
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
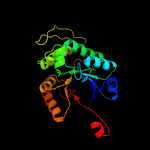
| 4 | d1luca_
|
|
 |
100.0 |
18 |
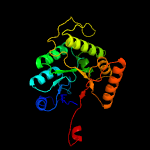
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
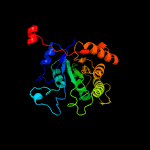
| 5 | c5w4zA_
|
|
 |
100.0 |
21 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
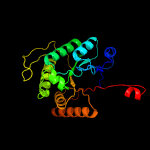
| 6 | c6friD_
|
|
 |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
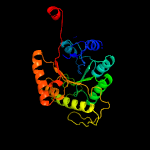
| 7 | c3sdoB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
| 8 | d1tvla_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 9 | c1tvlA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 10 | d1ezwa_
|
|
 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 11 | c3b9nB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 12 | c1z69D_
|
|
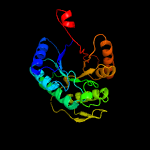 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
| 13 | c6ak1B_
|
|
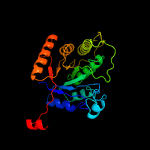 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
|
| 14 | c5dqpA_
|
|
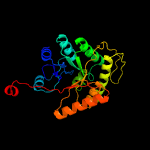 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 15 | c3raoB_
|
|
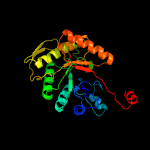 |
100.0 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
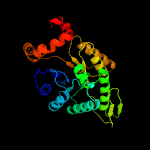
| 16 | c2wgkA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
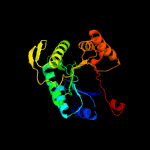
| 17 | c2i7gA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
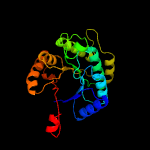
| 18 | d1f07a_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
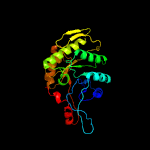
| 19 | d1rhca_
|
|
 |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 20 | c3c8nB_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 21 | c5wanA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
| 22 | d1nqka_ |
|
not modelled |
100.0 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
| 23 | d1nfpa_ |
|
not modelled |
99.7 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
97.6 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | c3dcpB_ |
|
not modelled |
59.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of the putative histidinol phosphatase hisk from2 listeria monocytogenes. northeast structural genomics consortium3 target lmr141.
|
|
|
| 26 | c4ovxA_ |
|
not modelled |
56.0 |
32 |
PDB header:isomerase
Chain: A: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structure of xylose isomerase domain protein from planctomyces2 limnophilus dsm 3776
|
|
|
| 27 | c3e38A_ |
|
not modelled |
53.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:two-domain protein containing predicted php-like metal-
PDBTitle: crystal structure of a two-domain protein containing predicted php-2 like metal-dependent phosphoesterase (bvu_3505) from bacteroides3 vulgatus atcc 8482 at 2.20 a resolution
|
|
|
| 28 | c3fa4D_ |
|
not modelled |
52.6 |
17 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 29 | c2yb1A_ |
|
not modelled |
52.4 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
|
|
|
| 30 | c1zlpA_ |
|
not modelled |
50.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 31 | d1uoua2 |
|
not modelled |
50.0 |
16 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 32 | c2zvrA_ |
|
not modelled |
49.5 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related protein from2 thermotoga maritima
|
|
|
| 33 | c2w9mB_ |
|
not modelled |
47.3 |
11 |
PDB header:dna replication
Chain: B: PDB Molecule:polymerase x;
PDBTitle: structure of family x dna polymerase from deinococcus2 radiodurans
|
|
|
| 34 | d1xp3a1 |
|
not modelled |
47.2 |
7 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
|
|
| 35 | d1m5wa_ |
|
not modelled |
46.2 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Pyridoxine 5'-phosphate synthase
Family:Pyridoxine 5'-phosphate synthase |
|
|
| 36 | c5dlcC_ |
|
not modelled |
46.0 |
33 |
PDB header:transferase
Chain: C: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
|
|
|
| 37 | c4lsbA_ |
|
not modelled |
45.6 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:lyase/mutase;
PDBTitle: crystal structure of a putative lyase/mutase from burkholderia2 cenocepacia j2315
|
|
|
| 38 | c3qy6A_ |
|
not modelled |
44.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 39 | c3d0cB_ |
|
not modelled |
44.4 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 40 | d1m65a_ |
|
not modelled |
41.2 |
10 |
Fold:7-stranded beta/alpha barrel
Superfamily:PHP domain-like
Family:PHP domain |
|
|
| 41 | c3e0fA_ |
|
not modelled |
39.7 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative metal-dependent phosphoesterase;
PDBTitle: crystal structure of a putative metal-dependent phosphoesterase2 (bad_1165) from bifidobacterium adolescentis atcc 15703 at 2.40 a3 resolution
|
|
|
| 42 | c3vniC_ |
|
not modelled |
38.4 |
11 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 43 | c2hjpA_ |
|
not modelled |
37.2 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
|
|
| 44 | c2yz5B_ |
|
not modelled |
37.1 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol phosphatase;
PDBTitle: histidinol phosphate phosphatase complexed with phosphate
|
|
|
| 45 | c2x7vA_ |
|
not modelled |
36.7 |
7 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
|
|
| 46 | c3ih1A_ |
|
not modelled |
36.7 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 47 | c3e96B_ |
|
not modelled |
36.4 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from bacillus2 clausii
|
|
|
| 48 | c2ze3A_ |
|
not modelled |
36.2 |
25 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 49 | d1thfd_ |
|
not modelled |
35.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 50 | c3gk0H_ |
|
not modelled |
35.7 |
18 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein from2 burkholderia pseudomallei
|
|
|
| 51 | c4gx9A_ |
|
not modelled |
35.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase iii subunit epsilon,dna polymerase iii
PDBTitle: crystal structure of a dna polymerase iii alpha-epsilon chimera
|
|
|
| 52 | c3lyeA_ |
|
not modelled |
34.6 |
47 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 53 | c4ur7B_ |
|
not modelled |
33.5 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:keto-deoxy-d-galactarate dehydratase;
PDBTitle: crystal structure of keto-deoxy-d-galactarate dehydratase2 complexed with pyruvate
|
|
|
| 54 | c6k0aC_ |
|
not modelled |
32.1 |
11 |
PDB header:rna binding protein/rna
Chain: C: PDB Molecule:ribonuclease p protein component 3;
PDBTitle: cryo-em structure of an archaeal ribonuclease p
|
|
|
| 55 | d1s2wa_ |
|
not modelled |
32.0 |
33 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 56 | c3b8iF_ |
|
not modelled |
31.2 |
12 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 57 | c5zhzA_ |
|
not modelled |
29.7 |
29 |
PDB header:dna binding protein
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of the apurinic/apyrimidinic endonuclease iv from2 mycobacterium tuberculosis
|
|
|
| 58 | d1m3ua_ |
|
not modelled |
27.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 59 | c5i4rA_ |
|
not modelled |
27.2 |
11 |
PDB header:toxin/antitoxin
Chain: A: PDB Molecule:contact-dependent inhibitor a;
PDBTitle: contact-dependent inhibition system from escherichia coli nc101 -2 ternary cdia/cdii/ef-tu complex (trypsin-modified)
|
|
|
| 60 | d1oy0a_ |
|
not modelled |
27.0 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 61 | d1brwa2 |
|
not modelled |
26.9 |
18 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 62 | c2wjeA_ |
|
not modelled |
26.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
|
|
| 63 | c3o6cA_ |
|
not modelled |
26.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: pyridoxal phosphate biosynthetic protein pdxj from campylobacter2 jejuni
|
|
|
| 64 | d1o5ka_ |
|
not modelled |
25.4 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 65 | c3auoB_ |
|
not modelled |
24.8 |
15 |
PDB header:transferase/dna
Chain: B: PDB Molecule:dna polymerase beta family (x family);
PDBTitle: dna polymerase x from thermus thermophilus hb8 ternary complex with 1-2 nt gapped dna and ddgtp
|
|
|
| 66 | c2y85D_ |
|
not modelled |
24.7 |
10 |
PDB header:isomerase
Chain: D: PDB Molecule:phosphoribosyl isomerase a;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphoribosyl2 isomerase with bound rcdrp
|
|
|
| 67 | c3obeB_ |
|
not modelled |
24.3 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
|
|
| 68 | d1ka9f_ |
|
not modelled |
24.2 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 69 | c3cnyA_ |
|
not modelled |
23.7 |
13 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
|
|
| 70 | c4cngB_ |
|
not modelled |
23.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:spou rrna methylase;
PDBTitle: crystal structure of sulfolobus acidocaldarius trmj in2 complex with s-adenosyl-l-homocysteine
|
|
|
| 71 | c2qiwA_ |
|
not modelled |
22.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 72 | c2anuA_ |
|
not modelled |
21.6 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:hypothetical protein tm0559;
PDBTitle: crystal structure of predicted metal-dependent phosphoesterase (php2 family) (tm0559) from thermotoga maritima at 2.40 a resolution
|
|
|
| 73 | d2anua1 |
|
not modelled |
21.6 |
15 |
Fold:7-stranded beta/alpha barrel
Superfamily:PHP domain-like
Family:PHP domain |
|
|
| 74 | d1muma_ |
|
not modelled |
21.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 75 | c3tdmD_ |
|
not modelled |
21.1 |
19 |
PDB header:de novo protein
Chain: D: PDB Molecule:computationally designed two-fold symmetric tim-barrel
PDBTitle: computationally designed tim-barrel protein, halfflr
|
|
|
| 76 | c4wd0A_ |
|
not modelled |
19.8 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of hisap form arthrobacter aurescens
|
|
|
| 77 | d2a6na1 |
|
not modelled |
19.7 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 78 | c3bh1A_ |
|
not modelled |
18.6 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
|
|
| 79 | c4gj1A_ |
|
not modelled |
17.5 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)2 methylideneamino] imidazole-4-carboxamide isomerase (hisa).
|
|
|
| 80 | c3dx5A_ |
|
not modelled |
15.9 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:uncharacterized protein asbf;
PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
|
|
|
| 81 | d1ujqa_ |
|
not modelled |
15.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 82 | c3eooL_ |
|
not modelled |
15.6 |
19 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 83 | d1xxxa1 |
|
not modelled |
14.8 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 84 | d1x7fa2 |
|
not modelled |
14.4 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
|
|
| 85 | c4mg4G_ |
|
not modelled |
14.3 |
17 |
PDB header:unknown function
Chain: G: PDB Molecule:phosphonomutase;
PDBTitle: crystal structure of a putative phosphonomutase from burkholderia2 cenocepacia j2315
|
|
|
| 86 | d1e8ca2 |
|
not modelled |
14.2 |
12 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
|
|
| 87 | c3ez4B_ |
|
not modelled |
14.1 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate hydroxymethyltransferase2 from burkholderia pseudomallei
|
|
|
| 88 | d1oyaa_ |
|
not modelled |
13.8 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 89 | c3orsD_ |
|
not modelled |
13.7 |
10 |
PDB header:isomerase,biosynthetic protein
Chain: D: PDB Molecule:n5-carboxyaminoimidazole ribonucleotide mutase;
PDBTitle: crystal structure of n5-carboxyaminoimidazole ribonucleotide mutase2 from staphylococcus aureus
|
|
|
| 90 | c3umuA_ |
|
not modelled |
13.5 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of l-histidinol phosphate phosphatase (hisk) from2 lactococcus lactis subsp. lactis il1403 complexed with zn, phosphate3 and l-histidinol
|
|
|
| 91 | d1z41a1 |
|
not modelled |
13.1 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 92 | c3i4eA_ |
|
not modelled |
13.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:isocitrate lyase;
PDBTitle: crystal structure of isocitrate lyase from burkholderia2 pseudomallei
|
|
|
| 93 | d1w3ia_ |
|
not modelled |
12.7 |
5 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 94 | c3f55A_ |
|
not modelled |
12.7 |
21 |
PDB header:hydrolase, allergen
Chain: A: PDB Molecule:beta-1,3-glucanase;
PDBTitle: crystal structure of the native endo beta-1,3-glucanase (hev b 2), a2 major allergen from hevea brasiliensis (space group p41)
|
|
|
| 95 | d1vzwa1 |
|
not modelled |
12.6 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Histidine biosynthesis enzymes |
|
|
| 96 | c6ahuJ_ |
|
not modelled |
12.3 |
7 |
PDB header:hydrolase/rna
Chain: J: PDB Molecule:ribonuclease p protein subunit p30;
PDBTitle: cryo-em structure of human ribonuclease p with mature trna
|
|
|
| 97 | d1viza_ |
|
not modelled |
12.0 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 98 | c3fkkA_ |
|
not modelled |
11.8 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 99 | c5aheA_ |
|
not modelled |
11.8 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:
PDBTitle: crystal structure of salmonella enterica hisa
|
|
|