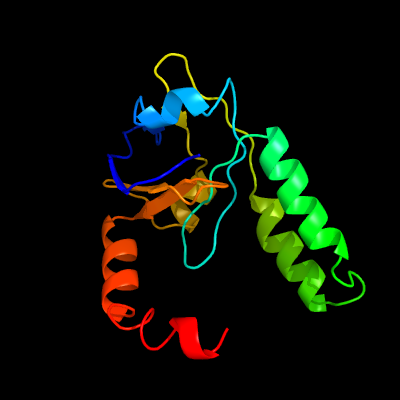
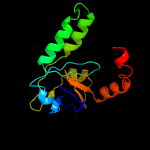
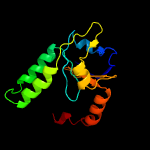
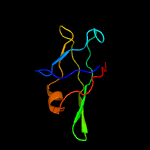
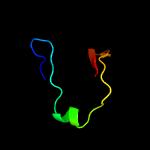
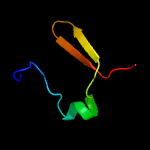
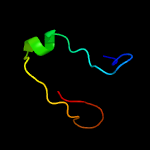
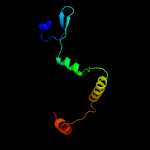
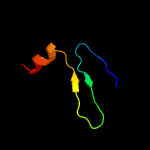
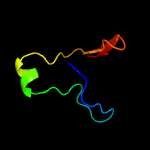
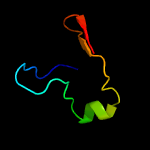
1 c1n0fF_
100.0
28
PDB header: biosynthetic proteinChain: F: PDB Molecule: protein mraz;PDBTitle: crystal structure of a cell division and cell wall2 biosynthesis protein upf0040 from mycoplasma pneumoniae:3 indication of a novel fold with a possible new conserved4 sequence motif
2 d1n0ea_
100.0
28
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: Hypothetical protein MraZ
3 c2glwA_
95.7
17
PDB header: transcriptionChain: A: PDB Molecule: 92aa long hypothetical protein;PDBTitle: the solution structure of phs018 from pyrococcus horikoshii
4 c2w1tB_
94.5
26
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: crystal structure of b. subtilis spovt
5 d1yfba1
90.5
22
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like
6 d2fy9a1
90.4
31
Fold: Double-split beta-barrelSuperfamily: AbrB/MazE/MraZ-likeFamily: AbrB N-terminal domain-like
7 c2ro5B_
88.2
26
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: rdc-refined solution structure of the n-terminal dna recognition2 domain of the bacillus subtilis transition-state regulator spovt
8 c3oeiB_
58.7
14
PDB header: toxin, protein bindingChain: B: PDB Molecule: relj (antitoxin rv3357);PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
9 c2l66B_
54.5
24
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
10 c2dgyA_
31.6
18
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
11 d2a6qb1
31.5
14
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
12 c3hs2H_
25.3
20
PDB header: antitoxinChain: H: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd truncated to residue 57 in an orthorhombic2 space group
13 d2a6qa1
23.6
14
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
14 c4mnoA_
23.0
18
PDB header: translationChain: A: PDB Molecule: translation initiation factor 1a;PDBTitle: crystal structure of aif1a from pyrococcus abyssi
15 c2k2eA_
22.8
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bp2786;PDBTitle: solution nmr structure of bordetella pertussis protein2 bp2786, a mth938-like domain. northeast structural3 genomics consortium target ber31
16 c3nnqA_
22.6
29
PDB header: viral proteinChain: A: PDB Molecule: n-terminal domain of moloney murine leukemia virusPDBTitle: crystal structure of the n-terminal domain of moloney murine leukemia2 virus integrase, northeast structural genomics consortium target or3
17 c3hryA_
22.3
21
PDB header: antitoxinChain: A: PDB Molecule: prevent host death protein;PDBTitle: crystal structure of phd in a trigonal space group and partially2 disordered
18 c3qq5A_
17.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: small gtp-binding protein;PDBTitle: crystal structure of the [fefe]-hydrogenase maturation protein hydf
19 c2c45F_
15.3
25
PDB header: lyaseChain: F: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: native precursor of pyruvoyl dependent aspartate decarboxylase
20 c1pt1B_
14.4
10
PDB header: lyaseChain: B: PDB Molecule: aspartate 1-decarboxylase;PDBTitle: unprocessed pyruvoyl dependent aspartate decarboxylase with histidine2 11 mutated to alanine
21 d2odka1
not modelled
14.1
21
Fold: YefM-likeSuperfamily: YefM-likeFamily: YefM-like
22 c5os9A_
not modelled
14.1
23
PDB header: dna binding proteinChain: A: PDB Molecule: b3 domain-containing transcription factor nga1;PDBTitle: structure of the b3 dna-binding domain of nga1
23 d1wida_
not modelled
14.0
23
Fold: DNA-binding pseudobarrel domainSuperfamily: DNA-binding pseudobarrel domainFamily: B3 DNA binding domain
24 c5l2dA_
not modelled
14.0
28
PDB header: cell adhesionChain: A: PDB Molecule: surface-associated protein csha;PDBTitle: streptococcal surface adhesin - csha nr2
25 c3g5oA_
not modelled
13.8
10
PDB header: toxin/antitoxinChain: A: PDB Molecule: uncharacterized protein rv2865;PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
26 d1s6la1
not modelled
13.8
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like
27 d2q4qa1
not modelled
13.8
14
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
28 d1ppya_
not modelled
13.2
10
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Pyruvoyl dependent aspartate decarboxylase, ADC
29 d2fi9a1
not modelled
13.2
15
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
30 c2zcpA_
not modelled
13.2
16
PDB header: transferaseChain: A: PDB Molecule: dehydrosqualene synthase;PDBTitle: crystal structure of the c(30) carotenoid dehydrosqualene synthase2 from staphylococcus aureus complexed with farnesyl thiopyrophosphate
31 c2gm2A_
not modelled
12.9
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: nmr structure of xanthomonas campestris xcc1710: northeast2 structural genomics consortium target xcr35
32 d2zgwa1
not modelled
12.9
56
Fold: SH3-like barrelSuperfamily: C-terminal domain of transcriptional repressorsFamily: Biotin repressor (BirA)
33 c3d55A_
not modelled
12.7
18
PDB header: toxin inhibitorChain: A: PDB Molecule: uncharacterized protein rv3357/mt3465;PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
34 c4hd1A_
not modelled
12.6
21
PDB header: transferaseChain: A: PDB Molecule: squalene synthase hpnc;PDBTitle: crystal structure of squalene synthase hpnc from alicyclobacillus2 acidocaldarius
35 c2odkD_
not modelled
12.4
21
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein;PDBTitle: putative prevent-host-death protein from nitrosomonas europaea
36 c2kdnA_
not modelled
12.2
7
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein pfe0790c;PDBTitle: solution structure of pfe0790c, a putative bola-like protein from the2 protozoan parasite plasmodium falciparum.
37 c5iysA_
not modelled
11.7
21
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: phytoene synthase;PDBTitle: crystal structure of a dehydrosqualene synthase in complex with ligand
38 c3dwmA_
not modelled
10.6
13
PDB header: transferaseChain: A: PDB Molecule: 9.5 kda culture filtrate antigen cfp10a;PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
39 c5l6mA_
not modelled
10.6
17
PDB header: hydrolaseChain: A: PDB Molecule: vapb family protein;PDBTitle: structure of caulobacter crescentus vapbc1 (vapb1deltac:vapc1 form)
40 d1d7qa_
not modelled
10.0
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
41 d1lpfa2
not modelled
10.0
23
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
42 c3df0C_
not modelled
10.0
27
PDB header: hydrolaseChain: C: PDB Molecule: calpastatin;PDBTitle: calcium-dependent complex between m-calpain and calpastatin
43 d2fvta1
not modelled
9.4
14
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
44 c4n6eB_
not modelled
9.1
14
PDB header: lyase/biosynthetic proteinChain: B: PDB Molecule: this/moad family protein;PDBTitle: crystal structure of amycolatopsis orientalis bexx/cyso complex
45 c6h3cC_
not modelled
8.9
17
PDB header: signaling proteinChain: C: PDB Molecule: brisc and brca1-a complex member 2;PDBTitle: cryo-em structure of the brisc complex bound to shmt2
46 c6h9xA_
not modelled
8.9
32
PDB header: ligaseChain: A: PDB Molecule: serine--trna ligase;PDBTitle: klebsiella pneumoniae seryl-trna synthetase in complex with the2 intermediate analog 5'-o-(n-(l-seryl)-sulfamoyl)adenosine
47 c3j81i_
not modelled
8.2
13
PDB header: ribosomeChain: I: PDB Molecule: es8;PDBTitle: cryoem structure of a partial yeast 48s preinitiation complex
48 c3errB_
not modelled
7.9
18
PDB header: ligaseChain: B: PDB Molecule: fusion protein of microtubule binding domain from mousePDBTitle: microtubule binding domain from mouse cytoplasmic dynein as a fusion2 with seryl-trna synthetase
49 c2oqkA_
not modelled
7.9
13
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
50 c3r5tA_
not modelled
7.8
17
PDB header: metal transportChain: A: PDB Molecule: ferric vibriobactin abc transporter, periplasmic ferricPDBTitle: crystal structure of holo-viup
51 c2dq3A_
not modelled
7.4
15
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of aq_298
52 d2gv8a2
not modelled
7.3
19
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
53 d1m7ja1
not modelled
7.0
23
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: D-aminoacylase
54 c3j39O_
not modelled
6.6
15
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l13a;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
55 d1xova1
not modelled
6.5
50
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: Ply C-terminal domain-like
56 d1v59a2
not modelled
6.5
20
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
57 c3if4C_
not modelled
6.5
31
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: integron cassette protein hfx_cass5;PDBTitle: structure from the mobile metagenome of north west arm sewage outfall:2 integron cassette protein hfx_cass5
58 c2ubpC_
not modelled
6.5
26
PDB header: hydrolaseChain: C: PDB Molecule: protein (urease alpha subunit);PDBTitle: structure of native urease from bacillus pasteurii
59 c2dq0A_
not modelled
6.3
12
PDB header: ligaseChain: A: PDB Molecule: seryl-trna synthetase;PDBTitle: crystal structure of seryl-trna synthetase from pyrococcus2 horikoshii complexed with a seryl-adenylate analog
60 c3qoqC_
not modelled
6.3
32
PDB header: transcription/dnaChain: C: PDB Molecule: alginate and motility regulator z;PDBTitle: crystal structure of the transcription factor amrz in complex with the2 18 base pair amrz1 binding site
61 d1d6za4
not modelled
6.2
10
Fold: N domain of copper amine oxidase-likeSuperfamily: Copper amine oxidase, domain NFamily: Copper amine oxidase, domain N
62 d1jt8a_
not modelled
6.2
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
63 c4e54B_
not modelled
6.1
21
PDB header: dna binding protein/dnaChain: B: PDB Molecule: dna damage-binding protein 2;PDBTitle: damaged dna induced uv-damaged dna-binding protein (uv-ddb)2 dimerization and its roles in chromatinized dna repair
64 c2mhdA_
not modelled
6.0
58
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr structure of the protein bacuni_03114 from bacteroides uniformis2 atcc 8492
65 d1seta2
not modelled
5.8
15
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain
66 c6bdcA_
not modelled
5.7
23
PDB header: protein transportChain: A: PDB Molecule: hcp1;PDBTitle: structure of hcp1 from flavobacterium johnsoniae
67 c3crcB_
not modelled
5.7
28
PDB header: hydrolaseChain: B: PDB Molecule: protein mazg;PDBTitle: crystal structure of escherichia coli mazg, the regulator2 of nutritional stress response
68 d1lvla2
not modelled
5.7
24
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
69 c1sryB_
not modelled
5.6
16
PDB header: ligase(synthetase)Chain: B: PDB Molecule: seryl-trna synthetase;PDBTitle: refined crystal structure of the seryl-trna synthetase from2 thermus thermophilus at 2.5 angstroms resolution
70 d2j7ja2
not modelled
5.6
30
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
71 c3zvkG_
not modelled
5.5
11
PDB header: antitoxin/toxin/dnaChain: G: PDB Molecule: antitoxin of toxin-antitoxin system vapb;PDBTitle: crystal structure of vapbc2 from rickettsia felis bound to2 a dna fragment from their promoter
72 d1ihna_
not modelled
5.5
9
Fold: MTH938-likeSuperfamily: MTH938-likeFamily: MTH938-like
73 c3vqzA_
not modelled
5.4
15
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: metallo-beta-lactamase;PDBTitle: crystal structure of metallo-beta-lactamase, smb-1, in a complex with2 mercaptoacetic acid
74 c3jywM_
not modelled
5.4
14
PDB header: ribosomeChain: M: PDB Molecule: 60s ribosomal protein l16(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
75 d1k1da1
not modelled
5.2
29
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)
76 c5k92C_
not modelled
5.1
22
PDB header: de novo proteinChain: C: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
77 c5k92B_
not modelled
5.1
22
PDB header: de novo proteinChain: B: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide
78 c5k92A_
not modelled
5.1
22
PDB header: de novo proteinChain: A: PDB Molecule: apo-(csl16c)3;PDBTitle: crystal structure of an apo tris-thiolate binding site in a de novo2 three stranded coiled coil peptide