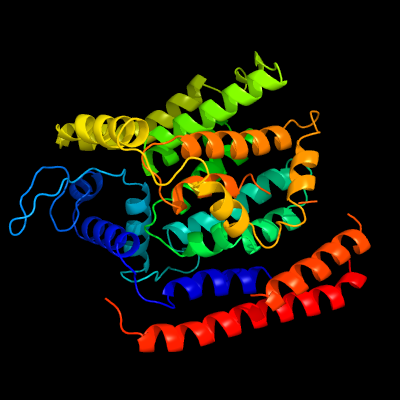
| 1 | c5f15A_
|
|
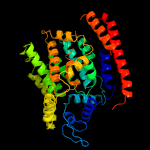 |
99.3 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
|
|
|
|
| 2 | c6p25A_
|
|
 |
98.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
|
|
|
|
| 3 | c6p2rB_
|
|
 |
98.3 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
|
|
|
|
| 4 | c3wajA_
|
|
 |
98.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
|
|
|
|
| 5 | c3rceA_
|
|
 |
95.8 |
10 |
PDB header:transferase/peptide
Chain: A: PDB Molecule:oligosaccharide transferase to n-glycosylate proteins;
PDBTitle: bacterial oligosaccharyltransferase pglb
|
|
|
|
| 6 | c6eznF_
|
|
 |
77.4 |
8 |
PDB header:membrane protein
Chain: F: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
|
|
|
|
| 7 | d1ynjd1
|
|
 |
20.2 |
44 |
Fold:beta and beta-prime subunits of DNA dependent RNA-polymerase
Superfamily:beta and beta-prime subunits of DNA dependent RNA-polymerase
Family:RNA-polymerase beta-prime |
|
|
|
| 8 | d1zxoa1
|
|
 |
16.4 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
|
| 9 | c3e1kD_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: D: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 10 | c3e1kF_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: F: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 11 | c3e1kP_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: P: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 12 | c3e1kB_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: B: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 13 | c3e1kL_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: L: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 14 | c3e1kJ_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: J: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 15 | c3e1kH_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: H: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 16 | c3e1kN_
|
|
 |
15.7 |
27 |
PDB header:transcription
Chain: N: PDB Molecule:lactose regulatory protein lac9;
PDBTitle: crystal structure of kluyveromyces lactis gal80p in complex with the2 acidic activation domain of gal4p
|
|
|
|
| 17 | c4g7oN_
|
|
 |
15.2 |
50 |
PDB header:transcription, transferase/dna
Chain: N: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex containing 2 nt of rna
|
|
|
|
| 18 | c5xj0D_
|
|
 |
15.1 |
50 |
PDB header:transferase/transcription
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: t. thermophilus rna polymerase holoenzyme bound with gp39 and gp76
|
|
|
|
| 19 | c3fmtF_
|
|
 |
13.9 |
44 |
PDB header:replication inhibitor/dna
Chain: F: PDB Molecule:protein seqa;
PDBTitle: crystal structure of seqa bound to dna
|
|
|
|
| 20 | c5x22D_
|
|
 |
13.5 |
50 |
PDB header:transferase/dna
Chain: D: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: crystal structure of thermus thermophilus transcription initiation2 complex with gpa and cmpcpp
|
|
|
|
| 21 | d1zbsa2 |
|
not modelled |
13.3 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
| 22 | d2r6gf1 |
|
not modelled |
12.9 |
14 |
Fold:MalF N-terminal region-like
Superfamily:MalF N-terminal region-like
Family:MalF N-terminal region-like |
|
|
| 23 | d1xrxa1 |
|
not modelled |
12.6 |
50 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:SeqA N-terminal domain-like |
|
|
| 24 | c1xrxD_ |
|
not modelled |
12.6 |
50 |
PDB header:replication inhibitor
Chain: D: PDB Molecule:seqa protein;
PDBTitle: crystal structure of a dna-binding protein
|
|
|
| 25 | c5z3dA_ |
|
not modelled |
12.3 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycoside hydrolase 15-related protein;
PDBTitle: glycosidase f290y
|
|
|
| 26 | c4y0lA_ |
|
not modelled |
12.2 |
33 |
PDB header:membrane protein
Chain: A: PDB Molecule:putative membrane protein mmpl11;
PDBTitle: mycobacterial membrane protein mmpl11d2
|
|
|
| 27 | c4rymA_ |
|
not modelled |
12.2 |
9 |
PDB header:membrane protein
Chain: A: PDB Molecule:integral membrane protein;
PDBTitle: crystal structure of bctspo iodo type1 monomer
|
|
|
| 28 | d2hu7a1 |
|
not modelled |
11.6 |
57 |
Fold:7-bladed beta-propeller
Superfamily:Peptidase/esterase 'gauge' domain
Family:Acylamino-acid-releasing enzyme, N-terminal donain |
|
|
| 29 | c2lzqA_ |
|
not modelled |
11.6 |
50 |
PDB header:viral protein
Chain: A: PDB Molecule:ns2 peptide;
PDBTitle: structure of ns2(32-57) gbvb protein
|
|
|
| 30 | c5l75F_ |
|
not modelled |
11.4 |
13 |
PDB header:transport protein
Chain: F: PDB Molecule:fig000988: predicted permease;
PDBTitle: a protein structure
|
|
|
| 31 | c1zzaA_ |
|
not modelled |
11.3 |
29 |
PDB header:membrane protein
Chain: A: PDB Molecule:stannin;
PDBTitle: solution nmr structure of the membrane protein stannin
|
|
|
| 32 | c6ig4B_ |
|
not modelled |
11.0 |
32 |
PDB header:transferase
Chain: B: PDB Molecule:phosphatidate cytidylyltransferase, mitochondrial;
PDBTitle: structure of mitochondrial cdp-dag synthase tam41, delta 74
|
|
|
| 33 | c3hf1B_ |
|
not modelled |
10.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ribonucleoside-diphosphate reductase subunit m2 b;
PDBTitle: crystal structure of human p53r2
|
|
|
| 34 | c2npmB_ |
|
not modelled |
10.7 |
22 |
PDB header:protein binding
Chain: B: PDB Molecule:14-3-3 domain containing protein;
PDBTitle: crystal structure of cryptosporidium parvum 14-3-3 protein in complex2 with peptide
|
|
|
| 35 | c3efzA_ |
|
not modelled |
10.7 |
33 |
PDB header:signaling protein
Chain: A: PDB Molecule:14-3-3 protein;
PDBTitle: crystal structure of a 14-3-3 protein from cryptosporidium parvum2 (cgd1_2980)
|
|
|
| 36 | d3efza1 |
|
not modelled |
10.7 |
33 |
Fold:alpha-alpha superhelix
Superfamily:14-3-3 protein
Family:14-3-3 protein |
|
|
| 37 | c5x3hA_ |
|
not modelled |
10.5 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: the y81g mutant of the ung crystal structure from nitratifractor2 salsuginis
|
|
|
| 38 | d1t33a2 |
|
not modelled |
10.5 |
18 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
|
|
| 39 | c5yi8B_ |
|
not modelled |
10.4 |
50 |
PDB header:cell cycle
Chain: B: PDB Molecule:pon peptide from partner of numb;
PDBTitle: crystal structure of drosophila numb ptb domain and pon peptide2 complex
|
|
|
| 40 | c2lioA_ |
|
not modelled |
10.4 |
24 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of bfr322 from bacteroides fragilis, northeast2 structural genomics consortium target bfr322
|
|
|
| 41 | d1q90d_ |
|
not modelled |
10.4 |
8 |
Fold:a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Superfamily:a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Family:a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
| 42 | c3qqcA_ |
|
not modelled |
10.0 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:dna-directed rna polymerase subunit b, dna-directed rna
PDBTitle: crystal structure of archaeal spt4/5 bound to the rnap clamp domain
|
|
|
| 43 | d1o9da_ |
|
not modelled |
10.0 |
44 |
Fold:alpha-alpha superhelix
Superfamily:14-3-3 protein
Family:14-3-3 protein |
|
|
| 44 | c3dwbA_ |
|
not modelled |
10.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:endothelin-converting enzyme 1;
PDBTitle: structure of human ece-1 complexed with phosphoramidon
|
|
|
| 45 | c2xsbA_ |
|
not modelled |
9.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:hyaluronoglucosaminidase;
PDBTitle: ogoga pugnac complex
|
|
|
| 46 | c2c1nA_ |
|
not modelled |
9.7 |
56 |
PDB header:signaling protein
Chain: A: PDB Molecule:14-3-3 protein zeta/delta;
PDBTitle: molecular basis for the recognition of phosphorylated and2 phosphoacetylated histone h3 by 14-3-3
|
|
|
| 47 | c5un8B_ |
|
not modelled |
9.7 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:protein o-glcnacase;
PDBTitle: crystal structure of human o-glcnacase in complex with glycopeptide2 p53
|
|
|
| 48 | c3tr7A_ |
|
not modelled |
9.6 |
80 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: structure of a uracil-dna glycosylase (ung) from coxiella burnetii
|
|
|
| 49 | c4iuwA_ |
|
not modelled |
9.5 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:neutral endopeptidase;
PDBTitle: crystal structure of pepo from lactobacillus rhamnosis hn001 (dr20)
|
|
|
| 50 | c2k8jX_ |
|
not modelled |
9.4 |
31 |
PDB header:viral protein
Chain: X: PDB Molecule:p7tm2;
PDBTitle: solution structure of hcv p7 tm2
|
|
|
| 51 | c2yf3F_ |
|
not modelled |
9.3 |
43 |
PDB header:hydrolase
Chain: F: PDB Molecule:mazg-like nucleoside triphosphate pyrophosphohydrolase;
PDBTitle: crystal structure of dr2231, the mazg-like protein from deinococcus2 radiodurans, complex with manganese
|
|
|
| 52 | d2o02a1 |
|
not modelled |
9.2 |
56 |
Fold:alpha-alpha superhelix
Superfamily:14-3-3 protein
Family:14-3-3 protein |
|
|
| 53 | c2rfpA_ |
|
not modelled |
9.2 |
36 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative ntp pyrophosphohydrolase;
PDBTitle: crystal structure of putative ntp pyrophosphohydrolase2 (yp_001813558.1) from exiguobacterium sibiricum 255-15 at 1.74 a3 resolution
|
|
|
| 54 | c3zf7m_ |
|
not modelled |
9.2 |
33 |
PDB header:ribosome
Chain: M: PDB Molecule:60s ribosomal protein l12, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 55 | c4uc2B_ |
|
not modelled |
9.1 |
15 |
PDB header:membrane protein
Chain: B: PDB Molecule:translocator protein tspo;
PDBTitle: crystal structure of translocator protein 18kda (tspo) from2 rhodobacter sphaeroides (a139t mutant) in p212121 space group
|
|
|
| 56 | c4a19Q_ |
|
not modelled |
9.1 |
50 |
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l36;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
|
|
|
| 57 | d2vbua1 |
|
not modelled |
9.1 |
39 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin kinase-like
Family:CTP-dependent riboflavin kinase-like |
|
|
| 58 | c6g90J_ |
|
not modelled |
9.1 |
18 |
PDB header:splicing
Chain: J: PDB Molecule:u1 small nuclear ribonucleoprotein component snu71;
PDBTitle: prespliceosome structure provides insight into spliceosome assembly2 and regulation (map a2)
|
|
|
| 59 | c2vuxB_ |
|
not modelled |
9.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ribonucleoside-diphosphate reductase subunit m2 b;
PDBTitle: human ribonucleotide reductase, subunit m2 b
|
|
|
| 60 | c6edjD_ |
|
not modelled |
8.9 |
60 |
PDB header:virus like particle
Chain: D: PDB Molecule:external core antigen;
PDBTitle: cryo-em structure of woodchuck hepatitis virus capsid
|
|
|
| 61 | c5t6oA_ |
|
not modelled |
8.9 |
28 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:poly-beta-hydroxybuterate polymerase;
PDBTitle: structure of the catalytic domain of the class i polyhydroxybutyrate2 synthase from cupriavidus necator
|
|
|
| 62 | d2o8pa1 |
|
not modelled |
8.9 |
25 |
Fold:alpha-alpha superhelix
Superfamily:14-3-3 protein
Family:14-3-3 protein |
|
|
| 63 | c2owrD_ |
|
not modelled |
8.8 |
80 |
PDB header:hydrolase
Chain: D: PDB Molecule:uracil-dna glycosylase;
PDBTitle: crystal structure of vaccinia virus uracil-dna glycosylase
|
|
|
| 64 | c3j3bi_ |
|
not modelled |
8.8 |
50 |
PDB header:ribosome
Chain: I: PDB Molecule:60s ribosomal protein l10-like;
PDBTitle: structure of the human 60s ribosomal proteins
|
|
|
| 65 | c6n7xH_ |
|
not modelled |
8.8 |
18 |
PDB header:rna binding protein/rna
Chain: H: PDB Molecule:u1 small nuclear ribonucleoprotein component snu71;
PDBTitle: s. cerevisiae u1 snrnp
|
|
|
| 66 | c5ireD_ |
|
not modelled |
8.8 |
22 |
PDB header:virus
Chain: D: PDB Molecule:m protein;
PDBTitle: the cryo-em structure of zika virus
|
|
|
| 67 | c3eyiB_ |
|
not modelled |
8.7 |
46 |
PDB header:dna binding protein/dna
Chain: B: PDB Molecule:z-dna-binding protein 1;
PDBTitle: the crystal structure of the second z-dna binding domain of2 human dai (zbp1) in complex with z-dna
|
|
|
| 68 | c5lc5c_ |
|
not modelled |
8.7 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,
PDBTitle: structure of mammalian respiratory complex i, class2
|
|
|
| 69 | c5ldwc_ |
|
not modelled |
8.7 |
19 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,
PDBTitle: structure of mammalian respiratory complex i, class1
|
|
|
| 70 | d1dmta_ |
|
not modelled |
8.7 |
24 |
Fold:Zincin-like
Superfamily:Metalloproteases ("zincins"), catalytic domain
Family:Neutral endopeptidase (neprilysin) |
|
|
| 71 | c5diyB_ |
|
not modelled |
8.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:hyaluronidase;
PDBTitle: thermobaculum terrenum o-glcnac hydrolase mutant - d120n
|
|
|
| 72 | c4b6ai_ |
|
not modelled |
8.5 |
67 |
PDB header:ribosome
Chain: I: PDB Molecule:60s ribosomal protein l10;
PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
|
|
|
| 73 | c2l4mA_ |
|
not modelled |
8.5 |
46 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of the zbeta domain of human dai and its binding2 modes to b- and z-dna
|
|
|
| 74 | c4dtfA_ |
|
not modelled |
8.5 |
21 |
PDB header:toxin
Chain: A: PDB Molecule:vgrg protein;
PDBTitle: structure of a vgrg vibrio cholerae toxin acd domain in complex with2 amp-pnp and mg++
|
|
|
| 75 | c2m67A_ |
|
not modelled |
8.5 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:merf;
PDBTitle: full-length mercury transporter protein merf in lipid bilayer2 membranes
|
|
|
| 76 | c3e6yB_ |
|
not modelled |
8.5 |
50 |
PDB header:signaling protein
Chain: B: PDB Molecule:14-3-3-like protein c;
PDBTitle: structure of 14-3-3 in complex with the differentiation-inducing agent2 cotylenin a
|
|
|
| 77 | c4w8kB_ |
|
not modelled |
8.4 |
21 |
PDB header:viral protein
Chain: B: PDB Molecule:cas1 protein;
PDBTitle: crystal structure of a putative cas1 enzyme from vibrio phage icp1
|
|
|
| 78 | d1tlha_ |
|
not modelled |
8.4 |
43 |
Fold:Anti-sigma factor AsiA
Superfamily:Anti-sigma factor AsiA
Family:Anti-sigma factor AsiA |
|
|
| 79 | c3aoiN_ |
|
not modelled |
8.3 |
50 |
PDB header:transcription, transferase/dna/rna
Chain: N: PDB Molecule:dna-directed rna polymerase subunit beta';
PDBTitle: rna polymerase-gfh1 complex (crystal type 2)
|
|
|
| 80 | c1ckwA_ |
|
not modelled |
8.3 |
57 |
PDB header:metal transport
Chain: A: PDB Molecule:protein (cystic fibrosis transmembrane
PDBTitle: cystic fibrosis transmembrane conductance regulator:2 solution structures of peptides based on the phe508 region,3 the most common site of disease-causing delta-f508 mutation
|
|
|
| 81 | c5x55A_ |
|
not modelled |
8.2 |
80 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable uracil-dna glycosylase;
PDBTitle: crystal structure of mimivirus uracil-dna glycosylase
|
|
|
| 82 | c5zjiH_ |
|
not modelled |
8.2 |
21 |
PDB header:membrane protein
Chain: H: PDB Molecule:photosystem i reaction center subunit vi, chloroplastic;
PDBTitle: structure of photosystem i supercomplex with light-harvesting2 complexes i and ii
|
|
|
| 83 | c5nn7A_ |
|
not modelled |
8.2 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: kshv uracil-dna glycosylase, apo form
|
|
|
| 84 | c2cukC_ |
|
not modelled |
8.2 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerate dehydrogenase/glyoxylate reductase;
PDBTitle: crystal structure of tt0316 protein from thermus thermophilus hb8
|
|
|
| 85 | c5oypD_ |
|
not modelled |
8.1 |
39 |
PDB header:virus
Chain: D: PDB Molecule:minor capsid protein micp;
PDBTitle: sacbrood virus of honeybee
|
|
|
| 86 | c5lsfD_ |
|
not modelled |
8.1 |
39 |
PDB header:virus
Chain: D: PDB Molecule:vp4;
PDBTitle: sacbrood honeybee virus
|
|
|
| 87 | c6egvD_ |
|
not modelled |
8.1 |
39 |
PDB header:virus
Chain: D: PDB Molecule:minor capsid protein micp;
PDBTitle: sacbrood virus of honeybee
|
|
|
| 88 | c6eh1D_ |
|
not modelled |
8.1 |
39 |
PDB header:virus
Chain: D: PDB Molecule:minor capsid protein micp;
PDBTitle: sacbrood virus of honeybee - expansion state ii
|
|
|
| 89 | c6egxD_ |
|
not modelled |
8.1 |
39 |
PDB header:virus
Chain: D: PDB Molecule:minor capsid protein micp;
PDBTitle: sacbrood virus of honeybee - expansion state i
|
|
|
| 90 | c1qfqB_ |
|
not modelled |
8.1 |
25 |
PDB header:transcription/rna
Chain: B: PDB Molecule:36-mer n-terminal peptide of the n protein;
PDBTitle: bacteriophage lambda n-protein-nutboxb-rna complex
|
|
|
| 91 | c6e6aB_ |
|
not modelled |
8.1 |
63 |
PDB header:protein binding
Chain: B: PDB Molecule:inclusion membrane protein a;
PDBTitle: triclinic crystal form of inca g144a point mutant
|
|
|
| 92 | d2cbia2 |
|
not modelled |
8.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:alpha-D-glucuronidase/Hyaluronidase catalytic domain |
|
|
| 93 | c4lzxB_ |
|
not modelled |
8.0 |
15 |
PDB header:metal binding protein
Chain: B: PDB Molecule:iq domain-containing protein g;
PDBTitle: complex of iqcg and ca2+-free cam
|
|
|
| 94 | c3pv9D_ |
|
not modelled |
8.0 |
11 |
PDB header:immune system
Chain: D: PDB Molecule:putative uncharacterized protein ph1245;
PDBTitle: structure of ph1245, a cas1 from pyrococcus horikoshii
|
|
|
| 95 | c2vy8A_ |
|
not modelled |
8.0 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:polymerase basic protein 2;
PDBTitle: the 627-domain from influenza a virus polymerase pb22 subunit with glu-627
|
|
|
| 96 | c5nv8A_ |
|
not modelled |
7.9 |
46 |
PDB header:transferase
Chain: A: PDB Molecule:ef-p arginine 32 rhamnosyl-transferase;
PDBTitle: structural basis for earp-mediated arginine glycosylation of2 translation elongation factor ef-p
|
|
|
| 97 | c2f5zK_ |
|
not modelled |
7.8 |
18 |
PDB header:oxidoreductase/protein binding
Chain: K: PDB Molecule:pyruvate dehydrogenase protein x component;
PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 (e3) complexed to the e3-binding domain of human e3-3 binding protein
|
|
|
| 98 | c3j39i_ |
|
not modelled |
7.8 |
50 |
PDB header:ribosome
Chain: I: PDB Molecule:60s ribosomal protein l10;
PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
|
|
|
| 99 | c6hq8B_ |
|
not modelled |
7.7 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-1,3-oligosaccharide phosphorylase;
PDBTitle: bacterial beta-1,3-oligosaccharide phosphorylase from gh149 with2 laminarihexaose bound at a surface site
|
|
|