| 1 |
|
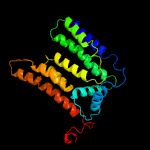
PDB 5f15 chain A
Region: 22 - 414
Aligned: 359
Modelled: 377
Confidence: 98.3%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
Phyre2
| 2 |
|
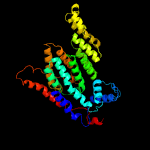
PDB 6p25 chain A
Region: 11 - 265
Aligned: 223
Modelled: 224
Confidence: 98.3%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
Phyre2
| 3 |
|
PDB 6p2r chain B
Region: 15 - 265
Aligned: 220
Modelled: 220
Confidence: 98.2%
Identity: 14%
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
Phyre2
| 4 |
|
PDB 3waj chain A
Region: 17 - 386
Aligned: 352
Modelled: 353
Confidence: 44.4%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
Phyre2
| 5 |
|
PDB 5c77 chain B
Region: 39 - 83
Aligned: 43
Modelled: 45
Confidence: 25.6%
Identity: 16%
PDB header:transferase
Chain: B: PDB Molecule:protein arginine n-methyltransferase sfm1;
PDBTitle: a novel protein arginine methyltransferase
Phyre2
| 6 |
|
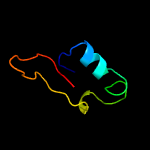
PDB 2zkr chain Y
Region: 1 - 16
Aligned: 16
Modelled: 16
Confidence: 9.0%
Identity: 38%
PDB header:ribosomal protein/rna
Chain: Y: PDB Molecule:5s ribosomal rna;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
Phyre2
| 7 |
|
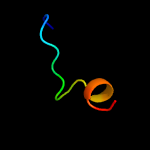
PDB 5d6h chain B
Region: 45 - 55
Aligned: 11
Modelled: 11
Confidence: 7.5%
Identity: 45%
PDB header:chaperone/protein transport
Chain: B: PDB Molecule:csua/b;
PDBTitle: crystal structure of csuc-csua/b chaperone-major subunit pre-assembly2 complex from csu biofilm-mediating pili of acinetobacter baumannii
Phyre2
| 8 |
|
PDB 3dwq chain D
Region: 47 - 89
Aligned: 32
Modelled: 43
Confidence: 7.0%
Identity: 34%
PDB header:toxin
Chain: D: PDB Molecule:subtilase cytotoxin, subunit b;
PDBTitle: crystal structure of the a-subunit of the ab5 toxin from e.2 coli with neu5gc-2,3gal-1,3glcnac
Phyre2
| 9 |
|
PDB 4k6l chain C
Region: 47 - 89
Aligned: 32
Modelled: 43
Confidence: 5.7%
Identity: 22%
PDB header:toxin
Chain: C: PDB Molecule:putative pertussis-like toxin subunit;
PDBTitle: structure of typhoid toxin
Phyre2
| 10 |
|
PDB 5whv chain F
Region: 47 - 89
Aligned: 32
Modelled: 43
Confidence: 5.6%
Identity: 25%
PDB header:toxin
Chain: F: PDB Molecule:artb protein;
PDBTitle: crystal structure of artb
Phyre2
| 11 |
|
PDB 2f69 chain A domain 1
Region: 59 - 83
Aligned: 25
Modelled: 25
Confidence: 5.6%
Identity: 28%
Fold: open-sided beta-meander
Superfamily: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain
Family: Histone H3 K4-specific methyltransferase SET7/9 N-terminal domain
Phyre2