| 1 |
|
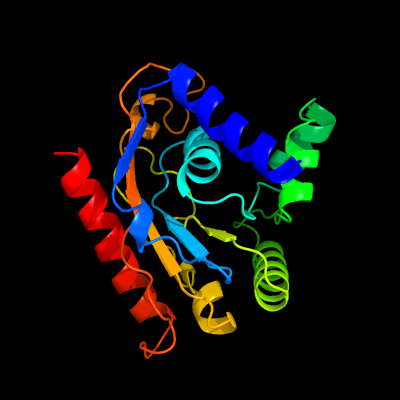
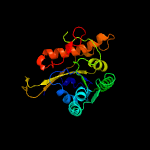
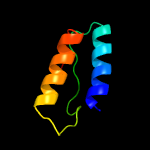
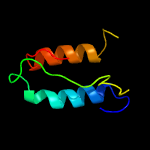
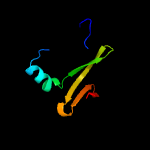
PDB 5kym chain A
Region: 2 - 206
Aligned: 193
Modelled: 205
Confidence: 100.0%
Identity: 29%
PDB header:transferase
Chain: A: PDB Molecule:1-acyl-sn-glycerol-3-phosphate acyltransferase;
PDBTitle: crystal structure of the 1-acyl-sn-glycerophosphate (lpa)2 acyltransferase, plsc, from thermotoga maritima
Phyre2
| 2 |
|
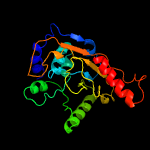
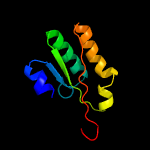
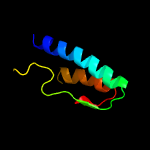
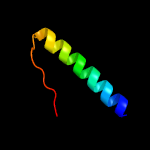
PDB 5f34 chain A
Region: 3 - 210
Aligned: 187
Modelled: 208
Confidence: 99.6%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol mannoside acyltransferase;
PDBTitle: crystal structure of membrane associated pata from mycobacterium2 smegmatis in complex with s-hexadecyl coenzyme a - p21 space group
Phyre2
| 3 |
|
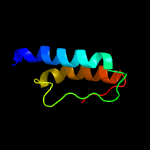
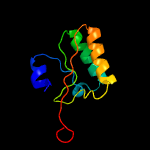
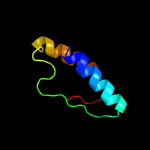
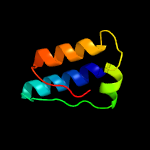
PDB 5knk chain B
Region: 21 - 229
Aligned: 183
Modelled: 209
Confidence: 99.5%
Identity: 13%
PDB header:transferase
Chain: B: PDB Molecule:lipid a biosynthesis lauroyl acyltransferase;
PDBTitle: lipid a secondary acyltransferase lpxm from acinetobacter baumannii2 with catalytic residue substitution (e127a)
Phyre2
| 4 |
|
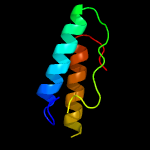
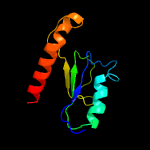
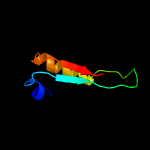
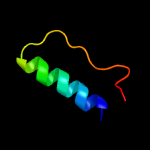
PDB 1iuq chain A
Region: 3 - 206
Aligned: 201
Modelled: 203
Confidence: 99.2%
Identity: 19%
Fold: Glycerol-3-phosphate (1)-acyltransferase
Superfamily: Glycerol-3-phosphate (1)-acyltransferase
Family: Glycerol-3-phosphate (1)-acyltransferase
Phyre2
| 5 |
|
PDB 5vrh chain A
Region: 90 - 147
Aligned: 58
Modelled: 58
Confidence: 23.9%
Identity: 14%
PDB header:transferase
Chain: A: PDB Molecule:apolipoprotein n-acyltransferase;
PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
Phyre2
| 6 |
|
PDB 3hkx chain A
Region: 88 - 147
Aligned: 60
Modelled: 60
Confidence: 16.7%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:amidase;
PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
Phyre2
| 7 |
|
PDB 4cyy chain A
Region: 95 - 147
Aligned: 53
Modelled: 53
Confidence: 15.7%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:pantetheinase;
PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
Phyre2
| 8 |
|
PDB 1yj5 chain B
Region: 76 - 158
Aligned: 77
Modelled: 83
Confidence: 14.9%
Identity: 18%
PDB header:transferase
Chain: B: PDB Molecule:5' polynucleotide kinase-3' phosphatase catalytic domain;
PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
Phyre2
| 9 |
|
PDB 3zvm chain A
Region: 76 - 158
Aligned: 77
Modelled: 83
Confidence: 13.3%
Identity: 18%
PDB header:hydrolase/transferase/dna
Chain: A: PDB Molecule:bifunctional polynucleotide phosphatase/kinase;
PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
Phyre2
| 10 |
|
PDB 1to6 chain A
Region: 110 - 205
Aligned: 91
Modelled: 96
Confidence: 11.9%
Identity: 20%
Fold: Glycerate kinase I
Superfamily: Glycerate kinase I
Family: Glycerate kinase I
Phyre2
| 11 |
|
PDB 2e11 chain B
Region: 89 - 147
Aligned: 59
Modelled: 59
Confidence: 9.7%
Identity: 14%
PDB header:hydrolase
Chain: B: PDB Molecule:hydrolase;
PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
Phyre2
| 12 |
|
PDB 4chm chain B
Region: 92 - 148
Aligned: 57
Modelled: 57
Confidence: 9.2%
Identity: 19%
PDB header:cell cycle
Chain: B: PDB Molecule:imc sub-compartment protein isp1;
PDBTitle: structure of inner membrane complex (imc) sub-compartment protein 12 (isp1) from toxoplasma gondii
Phyre2
| 13 |
|
PDB 2vhh chain A
Region: 88 - 147
Aligned: 60
Modelled: 60
Confidence: 9.1%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:cg3027-pa;
PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
Phyre2
| 14 |
|
PDB 5kha chain A
Region: 95 - 147
Aligned: 53
Modelled: 53
Confidence: 7.5%
Identity: 15%
PDB header:ligase
Chain: A: PDB Molecule:glutamine-dependent nad+ synthetase;
PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
Phyre2
| 15 |
|
PDB 3cwc chain B
Region: 105 - 150
Aligned: 46
Modelled: 46
Confidence: 7.5%
Identity: 24%
PDB header:transferase
Chain: B: PDB Molecule:putative glycerate kinase 2;
PDBTitle: crystal structure of putative glycerate kinase 2 from salmonella2 typhimurium lt2
Phyre2
| 16 |
|
PDB 4chj chain A
Region: 87 - 148
Aligned: 62
Modelled: 62
Confidence: 7.3%
Identity: 13%
PDB header:cell cycle
Chain: A: PDB Molecule:imc sub-compartment protein isp3;
PDBTitle: structure of inner membrane complex (imc) sub-compartment2 protein 3 (isp3) from toxoplasma gondii
Phyre2
| 17 |
|
PDB 6ftq chain A
Region: 88 - 119
Aligned: 32
Modelled: 32
Confidence: 6.5%
Identity: 13%
PDB header:hydrolase
Chain: A: PDB Molecule:beta-ureidopropionase;
PDBTitle: crystal structure of human beta-ureidopropionase (beta-alanine2 synthase) - mutant t299c
Phyre2
| 18 |
|
PDB 2zo7 chain A
Region: 16 - 35
Aligned: 18
Modelled: 20
Confidence: 6.3%
Identity: 28%
PDB header:luminescent protein
Chain: A: PDB Molecule:cyan/green-emitting gfp-like protein, kusabira-cyan mutant
PDBTitle: crystal structure of a kusabira-cyan mutant (kcy-r1), a cyan/green-2 emitting gfp-like protein
Phyre2
| 19 |
|
PDB 1ems chain B
Region: 95 - 148
Aligned: 54
Modelled: 54
Confidence: 6.2%
Identity: 11%
PDB header:antitumor protein
Chain: B: PDB Molecule:nit-fragile histidine triad fusion protein;
PDBTitle: crystal structure of the c. elegans nitfhit protein
Phyre2
| 20 |
|
PDB 3wuy chain A
Region: 95 - 123
Aligned: 29
Modelled: 29
Confidence: 5.9%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:nitrilase;
PDBTitle: crystal structure of nit6803
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|