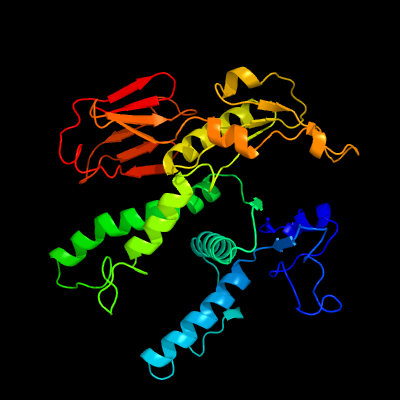
| 1 | c3igfB_
|
|
 |
100.0 |
25 |
PDB header:atp binding protein
Chain: B: PDB Molecule:all4481 protein;
PDBTitle: crystal structure of the all4481 protein from nostoc sp. pcc 7120,2 northeast structural genomics consortium target nsr300
|
|
|
|
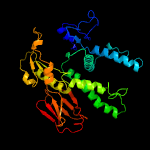
| 2 | c5zmfA_
|
|
 |
100.0 |
21 |
PDB header:hydrolase/transport protein
Chain: A: PDB Molecule:atpase arsa1;
PDBTitle: amppnp complex of c. reinhardtii arsa1
|
|
|
|
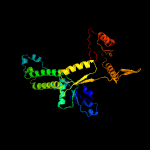
| 3 | c1ii0A_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:arsenical pump-driving atpase;
PDBTitle: crystal structure of the escherichia coli arsenite-translocating2 atpase
|
|
|
|
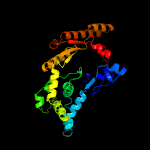
| 4 | c6bs5B_
|
|
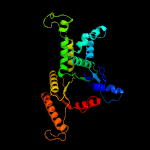 |
100.0 |
23 |
PDB header:unknown function
Chain: B: PDB Molecule:anion transporter;
PDBTitle: crystal structure of amp-pnp-bound bacterial get3-like a and b in2 mycobacterium tuberculosis
|
|
|
|
| 5 | c3ug7D_
|
|
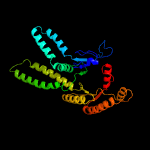 |
100.0 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:arsenical pump-driving atpase;
PDBTitle: crystal structure of get3 from methanocaldococcus jannaschii
|
|
|
|
| 6 | c3zq6D_
|
|
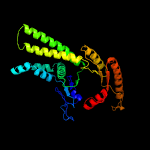 |
100.0 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative arsenical pump-driving atpase;
PDBTitle: adp-alf4 complex of m. therm. trc40
|
|
|
|
| 7 | c2wooC_
|
|
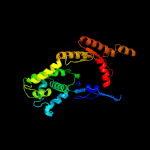 |
100.0 |
22 |
PDB header:hydrolase
Chain: C: PDB Molecule:atpase get3;
PDBTitle: nucleotide-free form of s. pombe get3
|
|
|
|
| 8 | c6bs3A_
|
|
 |
100.0 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:putative atpase rv3679;
PDBTitle: crystal structure of adp-bound bacterial get3-like a and b in2 mycobacterium tuberculosis
|
|
|
|
| 9 | c3ibgF_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: F: PDB Molecule:atpase, subunit of the get complex;
PDBTitle: crystal structure of aspergillus fumigatus get3 with bound2 adp
|
|
|
|
| 10 | c2wojD_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: D: PDB Molecule:atpase get3;
PDBTitle: adp-alf4 complex of s. cerevisiae get3
|
|
|
|
| 11 | c5bwkA_
|
|
 |
100.0 |
27 |
PDB header:hydrolase/transport
Chain: A: PDB Molecule:atpase get3;
PDBTitle: 6.0 a crystal structure of a get3-get4-get5 intermediate complex from2 s.cerevisiae
|
|
|
|
| 12 | d1ihua1
|
|
 |
100.0 |
25 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
|
| 13 | d1ihua2
|
|
 |
99.8 |
26 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
|
| 14 | c3io3A_
|
|
 |
99.8 |
29 |
PDB header:chaperone
Chain: A: PDB Molecule:deha2d07832p;
PDBTitle: get3 with adp from d. hansenii in closed form
|
|
|
|
| 15 | c3kjgB_
|
|
 |
98.3 |
20 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase complex, accessory
PDBTitle: adp-bound state of cooc1
|
|
|
|
| 16 | c4rzkA_
|
|
 |
98.3 |
10 |
PDB header:chaperone
Chain: A: PDB Molecule:small heat shock protein hsp20 family;
PDBTitle: crystal structure of sulfolobus solfataricus hsp20.1 acd
|
|
|
|
| 17 | c5mb8L_
|
|
 |
98.2 |
16 |
PDB header:chaperone
Chain: L: PDB Molecule:25.3 kda heat shock protein, chloroplastic;
PDBTitle: hsp21 dodecamer, structural model based on cryo-em and homology2 modelling
|
|
|
|
| 18 | c4zjdF_
|
|
 |
98.2 |
19 |
PDB header:chaperone
Chain: F: PDB Molecule:aggregation suppressing protein;
PDBTitle: small heat shock protein agsa from salmonella typhimurium: truncations2 at n- and c- termini
|
|
|
|
| 19 | c3w1zA_
|
|
 |
98.2 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 16;
PDBTitle: heat shock protein 16.0 from schizosaccharomyces pombe
|
|
|
|
| 20 | c3glaA_
|
|
 |
98.2 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:low molecular weight heat shock protein;
PDBTitle: crystal structure of the hspa from xanthomonas axonopodis
|
|
|
|
| 21 | c3w1zD_ |
|
not modelled |
98.2 |
14 |
PDB header:chaperone
Chain: D: PDB Molecule:heat shock protein 16;
PDBTitle: heat shock protein 16.0 from schizosaccharomyces pombe
|
|
|
| 22 | d1shsa_ |
|
not modelled |
98.1 |
17 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
|
|
| 23 | c1shsD_ |
|
not modelled |
98.1 |
17 |
PDB header:heat shock protein
Chain: D: PDB Molecule:small heat shock protein;
PDBTitle: small heat shock protein from methanococcus jannaschii
|
|
|
| 24 | c5mb8J_ |
|
not modelled |
98.1 |
15 |
PDB header:chaperone
Chain: J: PDB Molecule:25.3 kda heat shock protein, chloroplastic;
PDBTitle: hsp21 dodecamer, structural model based on cryo-em and homology2 modelling
|
|
|
| 25 | c4feiA_ |
|
not modelled |
98.1 |
34 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein-related protein;
PDBTitle: hsp17.7 from deinococcus radiodurans
|
|
|
| 26 | d1gmea_ |
|
not modelled |
98.1 |
20 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
|
|
| 27 | d2h50a1 |
|
not modelled |
98.0 |
28 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
|
|
| 28 | c4pbdA_ |
|
not modelled |
98.0 |
12 |
PDB header:protein binding
Chain: A: PDB Molecule:protein shq1 homolog;
PDBTitle: crystal structure of the n-terminal cs domain of human shq1
|
|
|
| 29 | c5ds1B_ |
|
not modelled |
97.9 |
20 |
PDB header:chaperone
Chain: B: PDB Molecule:17.1 kda class ii heat shock protein;
PDBTitle: core domain of the class ii small heat-shock protein hsp 17.7 from2 pisum sativum
|
|
|
| 30 | c2ygdV_ |
|
not modelled |
97.9 |
19 |
PDB header:chaperone
Chain: V: PDB Molecule:
PDBTitle: molecular architectures of the 24meric eye lens chaperone alphab-2 crystallin elucidated by a triple hybrid approach
|
|
|
| 31 | c3aabA_ |
|
not modelled |
97.9 |
13 |
PDB header:chaperone
Chain: A: PDB Molecule:putative uncharacterized protein st1653;
PDBTitle: small heat shock protein hsp14.0 with the mutations of i120f and i122f2 in the form i crystal
|
|
|
| 32 | d1gmeb_ |
|
not modelled |
97.9 |
23 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:HSP20 |
|
|
| 33 | c2wj5A_ |
|
not modelled |
97.8 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein beta-6;
PDBTitle: rat alpha crystallin domain
|
|
|
| 34 | c5ltwK_ |
|
not modelled |
97.8 |
21 |
PDB header:protein binding
Chain: K: PDB Molecule:heat shock protein beta-6;
PDBTitle: complex of human 14-3-3 sigma clu1 mutant with phosphorylated heat2 shock protein b6
|
|
|
| 35 | c2bolA_ |
|
not modelled |
97.8 |
20 |
PDB header:heat shock protein
Chain: A: PDB Molecule:small heat shock protein;
PDBTitle: crystal structure and assembly of tsp36, a metazoan small heat shock2 protein
|
|
|
| 36 | c2n3jA_ |
|
not modelled |
97.8 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein beta-1;
PDBTitle: solution structure of the alpha-crystallin domain from the redox-2 sensitive chaperone, hspb1
|
|
|
| 37 | c4ydzA_ |
|
not modelled |
97.8 |
14 |
PDB header:chaperone
Chain: A: PDB Molecule:stress-induced protein 1;
PDBTitle: stress-induced protein 1 from caenorhabditis elegans
|
|
|
| 38 | c3l1eA_ |
|
not modelled |
97.7 |
19 |
PDB header:chaperone
Chain: A: PDB Molecule:alpha-crystallin a chain;
PDBTitle: bovine alphaa crystallin zinc bound
|
|
|
| 39 | c4ylcF_ |
|
not modelled |
97.6 |
12 |
PDB header:chaperone
Chain: F: PDB Molecule:heat shock protein hsp20;
PDBTitle: crystal structure of del-c4 mutant of hsp14.1 from sulfolobus2 solfatataricus p2
|
|
|
| 40 | c3q9qB_ |
|
not modelled |
97.6 |
27 |
PDB header:chaperone
Chain: B: PDB Molecule:heat shock protein beta-1;
PDBTitle: hspb1 fragment second crystal form
|
|
|
| 41 | c6ewnA_ |
|
not modelled |
97.6 |
20 |
PDB header:chaperone
Chain: A: PDB Molecule:hspa;
PDBTitle: hspa from thermosynechococcus vulcanus in the presence of 2m urea with2 initial stages of denaturation
|
|
|
| 42 | c6f2rK_ |
|
not modelled |
97.6 |
17 |
PDB header:chaperone
Chain: K: PDB Molecule:hspb2,heat shock protein beta-2,heat shock protein beta-2,
PDBTitle: a hetrotetramer of human hspb2 and hspb3
|
|
|
| 43 | c3la6P_ |
|
not modelled |
97.5 |
19 |
PDB header:transferase
Chain: P: PDB Molecule:tyrosine-protein kinase wzc;
PDBTitle: octameric kinase domain of the e. coli tyrosine kinase wzc with bound2 adp
|
|
|
| 44 | c6dv5T_ |
|
not modelled |
97.5 |
18 |
PDB header:chaperone
Chain: T: PDB Molecule:heat shock protein beta-1;
PDBTitle: oligomeric complex of a hsp27 24-mer at 3.6 a resolution
|
|
|
| 45 | c2wj7D_ |
|
not modelled |
97.4 |
23 |
PDB header:chaperone
Chain: D: PDB Molecule:alpha-crystallin b chain;
PDBTitle: human alphab crystallin
|
|
|
| 46 | c2klrA_ |
|
not modelled |
97.4 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:alpha-crystallin b chain;
PDBTitle: solid-state nmr structure of the alpha-crystallin domain in alphab-2 crystallin oligomers
|
|
|
| 47 | c3cioA_ |
|
not modelled |
97.4 |
21 |
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:tyrosine-protein kinase etk;
PDBTitle: the kinase domain of escherichia coli tyrosine kinase etk
|
|
|
| 48 | c6f2rI_ |
|
not modelled |
97.4 |
21 |
PDB header:chaperone
Chain: I: PDB Molecule:hspb2,heat shock protein beta-2,heat shock protein beta-2,
PDBTitle: a hetrotetramer of human hspb2 and hspb3
|
|
|
| 49 | c6f2rE_ |
|
not modelled |
97.3 |
21 |
PDB header:chaperone
Chain: E: PDB Molecule:hspb2,heat shock protein beta-2,heat shock protein beta-2,
PDBTitle: a hetrotetramer of human hspb2 and hspb3
|
|
|
| 50 | c6f2rT_ |
|
not modelled |
97.3 |
26 |
PDB header:chaperone
Chain: T: PDB Molecule:heat shock protein beta-3,heat shock protein beta-3,heat
PDBTitle: a hetrotetramer of human hspb2 and hspb3
|
|
|
| 51 | c6nonB_ |
|
not modelled |
97.2 |
13 |
PDB header:dna binding protein
Chain: B: PDB Molecule:cobyrinic acid ac-diamide synthase;
PDBTitle: structure of cyanthece apo mcda
|
|
|
| 52 | c6g2gA_ |
|
not modelled |
97.2 |
25 |
PDB header:cytosolic protein
Chain: A: PDB Molecule:cytosolic fe-s cluster assembly factor cfd1;
PDBTitle: fe-s assembly cfd1
|
|
|
| 53 | c2qmoA_ |
|
not modelled |
97.2 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of dethiobiotin synthetase (biod) from helicobacter2 pylori
|
|
|
| 54 | c6iucC_ |
|
not modelled |
97.1 |
17 |
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:spooj regulator (soj);
PDBTitle: structure of helicobacter pylori soj-atp complex bound to dna
|
|
|
| 55 | c6f2rA_ |
|
not modelled |
97.0 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:hspb2,heat shock protein beta-2,heat shock protein beta-2,
PDBTitle: a hetrotetramer of human hspb2 and hspb3
|
|
|
| 56 | c4pfsA_ |
|
not modelled |
96.9 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:cobyrinic acid a,c-diamide synthase;
PDBTitle: crystal structure of cobyrinic acid a,c-diamide synthase from2 mycobacterium smegmatis
|
|
|
| 57 | c6gxzD_ |
|
not modelled |
96.8 |
16 |
PDB header:chaperone
Chain: D: PDB Molecule:pih1 domain-containing protein 1;
PDBTitle: crystal structure of the human rpap3(tpr2)-pih1d1(cs) complex
|
|
|
| 58 | c5zulB_ |
|
not modelled |
96.7 |
22 |
PDB header:chaperone
Chain: B: PDB Molecule:small heat shock protein;
PDBTitle: small heat shock protein from mycobacterium marinum m : form-3
|
|
|
| 59 | c3fmfA_ |
|
not modelled |
96.7 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of mycobacterium tuberculosis dethiobiotin2 synthetase complexed with 7,8 diaminopelargonic acid carbamate
|
|
|
| 60 | c5j1jA_ |
|
not modelled |
96.7 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:site-determining protein;
PDBTitle: structure of flen-amppnp complex
|
|
|
| 61 | c2vedA_ |
|
not modelled |
96.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:membrane protein capa1, protein tyrosine kinase;
PDBTitle: crystal structure of the chimerical mutant capabk55m2 protein
|
|
|
| 62 | c2ph1A_ |
|
not modelled |
96.6 |
17 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:nucleotide-binding protein;
PDBTitle: crystal structure of nucleotide-binding protein af2382 from2 archaeoglobus fulgidus, northeast structural genomics target gr165
|
|
|
| 63 | c2ozeA_ |
|
not modelled |
96.6 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:orf delta';
PDBTitle: the crystal structure of delta protein of psm19035 from2 streptoccocus pyogenes
|
|
|
| 64 | c3pg5A_ |
|
not modelled |
96.5 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein dip2308 from corynebacterium diphtheriae,2 northeast structural genomics consortium target cdr78
|
|
|
| 65 | c3endA_ |
|
not modelled |
96.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase
PDBTitle: crystal structure of the l protein of rhodobacter2 sphaeroides light-independent protochlorophyllide3 reductase (bchl) with mgadp bound: a homologue of the4 nitrogenase fe protein
|
|
|
| 66 | c3fkqA_ |
|
not modelled |
96.4 |
26 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ntrc-like two-domain protein;
PDBTitle: crystal structure of ntrc-like two-domain protein (rer070207001320)2 from eubacterium rectale at 2.10 a resolution
|
|
|
| 67 | c3k9gA_ |
|
not modelled |
96.4 |
25 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:pf-32 protein;
PDBTitle: crystal structure of a plasmid partition protein from borrelia2 burgdorferi at 2.25a resolution, iodide soak
|
|
|
| 68 | d1g3qa_ |
|
not modelled |
96.3 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 69 | c3vx3A_ |
|
not modelled |
96.3 |
25 |
PDB header:adp binding protein
Chain: A: PDB Molecule:atpase involved in chromosome partitioning, para/mind
PDBTitle: crystal structure of [nife] hydrogenase maturation protein hypb from2 thermococcus kodakarensis kod1
|
|
|
| 70 | c3ez6B_ |
|
not modelled |
96.2 |
21 |
PDB header:dna binding protein
Chain: B: PDB Molecule:plasmid partition protein a;
PDBTitle: structure of para-adp complex:tetragonal form
|
|
|
| 71 | d1iona_ |
|
not modelled |
96.2 |
17 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 72 | c4v02B_ |
|
not modelled |
96.0 |
22 |
PDB header:cell cycle
Chain: B: PDB Molecule:site-determining protein;
PDBTitle: minc:mind cell division protein complex, aquifex aeolicus
|
|
|
| 73 | d2afhe1 |
|
not modelled |
95.9 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 74 | c3ea0B_ |
|
not modelled |
95.7 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:atpase, para family;
PDBTitle: crystal structure of para family atpase from chlorobium tepidum tls
|
|
|
| 75 | c4rz3B_ |
|
not modelled |
95.6 |
26 |
PDB header:structural protein
Chain: B: PDB Molecule:site-determining protein;
PDBTitle: crystal structure of the mind-like atpase flhg
|
|
|
| 76 | c4ru8C_ |
|
not modelled |
95.3 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: structure of pnob8 para with amppnp
|
|
|
| 77 | c3q9lB_ |
|
not modelled |
94.9 |
39 |
PDB header:cell cycle, hydrolase
Chain: B: PDB Molecule:septum site-determining protein mind;
PDBTitle: the structure of the dimeric e.coli mind-atp complex
|
|
|
| 78 | d1byia_ |
|
not modelled |
94.8 |
21 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 79 | c3of5A_ |
|
not modelled |
94.7 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of a dethiobiotin synthetase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 80 | d1cp2a_ |
|
not modelled |
94.6 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 81 | c3ezfA_ |
|
not modelled |
93.0 |
38 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:para;
PDBTitle: partition protein
|
|
|
| 82 | c2bekB_ |
|
not modelled |
93.0 |
22 |
PDB header:chromosome segregation
Chain: B: PDB Molecule:segregation protein;
PDBTitle: structure of the bacterial chromosome segregation protein soj
|
|
|
| 83 | c3cwqB_ |
|
not modelled |
92.9 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:para family chromosome partitioning protein;
PDBTitle: crystal structure of chromosome partitioning protein (para) in complex2 with adp from synechocystis sp. northeast structural genomics3 consortium target sgr89
|
|
|
| 84 | c1hyqA_ |
|
not modelled |
92.6 |
18 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division inhibitor (mind-1);
PDBTitle: mind bacterial cell division regulator from a. fulgidus
|
|
|
| 85 | d1hyqa_ |
|
not modelled |
92.6 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 86 | c4dzzB_ |
|
not modelled |
91.4 |
13 |
PDB header:unknown function
Chain: B: PDB Molecule:plasmid partitioning protein parf;
PDBTitle: structure of parf-adp, crystal form 1
|
|
|
| 87 | c2og2A_ |
|
not modelled |
86.0 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:putative signal recognition particle receptor;
PDBTitle: crystal structure of chloroplast ftsy from arabidopsis2 thaliana
|
|
|
| 88 | c5l3qB_ |
|
not modelled |
85.5 |
22 |
PDB header:protein transport
Chain: B: PDB Molecule:signal recognition particle receptor subunit alpha;
PDBTitle: structure of the gtpase heterodimer of human srp54 and sralpha
|
|
|
| 89 | c3b9qA_ |
|
not modelled |
85.0 |
20 |
PDB header:protein transport
Chain: A: PDB Molecule:chloroplast srp receptor homolog, alpha subunit
PDBTitle: the crystal structure of cpftsy from arabidopsis thaliana
|
|
|
| 90 | c4a0rB_ |
|
not modelled |
80.1 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate
PDBTitle: structure of bifunctional dapa aminotransferase-dtb synthetase from2 arabidopsis thaliana bound to dethiobiotin (dtb).
|
|
|
| 91 | c4a0gC_ |
|
not modelled |
77.6 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:adenosylmethionine-8-amino-7-oxononanoate
PDBTitle: structure of bifunctional dapa aminotransferase-dtb synthetase from2 arabidopsis thaliana in its apo form.
|
|
|
| 92 | c2cnwF_ |
|
not modelled |
76.7 |
23 |
PDB header:signal recognition
Chain: F: PDB Molecule:cell division protein ftsy;
PDBTitle: gdpalf4 complex of the srp gtpases ffh and ftsy
|
|
|
| 93 | c2qy9A_ |
|
not modelled |
75.5 |
22 |
PDB header:protein transport
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
|
|
|
| 94 | c2yhsA_ |
|
not modelled |
72.4 |
24 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of the e. coli srp receptor ftsy
|
|
|
| 95 | c2xj9B_ |
|
not modelled |
70.7 |
29 |
PDB header:replication
Chain: B: PDB Molecule:mipz;
PDBTitle: dimer structure of the bacterial cell division regulator mipz
|
|
|
| 96 | c2iy3A_ |
|
not modelled |
70.2 |
24 |
PDB header:rna-binding
Chain: A: PDB Molecule:signal recognition particle protein,signal recognition
PDBTitle: structure of the e. coli signal regognition particle
|
|
|
| 97 | c1qzwC_ |
|
not modelled |
70.1 |
15 |
PDB header:signaling protein/rna
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
|
|
|
| 98 | c5l3rC_ |
|
not modelled |
68.3 |
10 |
PDB header:protein transport
Chain: C: PDB Molecule:signal recognition particle 54 kda protein, chloroplastic;
PDBTitle: structure of the gtpase heterodimer of chloroplast srp54 and ftsy from2 arabidopsis thaliana
|
|
|
| 99 | c2j7pA_ |
|
not modelled |
64.4 |
30 |
PDB header:signal recognition
Chain: A: PDB Molecule:signal recognition particle protein;
PDBTitle: gmppnp-stabilized ng domain complex of the srp gtpases ffh2 and ftsy
|
|
|
| 100 | c2q9cA_ |
|
not modelled |
63.4 |
21 |
PDB header:signaling protein
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of ftsy:gmppnp with mgcl complex
|
|
|
| 101 | c3dm5A_ |
|
not modelled |
61.8 |
21 |
PDB header:rna binding protein, transport protein
Chain: A: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: structures of srp54 and srp19, the two proteins assembling the2 ribonucleic core of the signal recognition particle from the archaeon3 pyrococcus furiosus.
|
|
|
| 102 | c5l3sF_ |
|
not modelled |
60.5 |
16 |
PDB header:protein transport
Chain: F: PDB Molecule:signal recognition particle receptor ftsy;
PDBTitle: structure of the gtpase heterodimer of crenarchaeal srp54 and ftsy
|
|
|
| 103 | c2j37W_ |
|
not modelled |
58.3 |
21 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein (srp54);
PDBTitle: model of mammalian srp bound to 80s rncs
|
|
|
| 104 | c1zu4A_ |
|
not modelled |
56.6 |
15 |
PDB header:protein transport
Chain: A: PDB Molecule:ftsy;
PDBTitle: crystal structure of ftsy from mycoplasma mycoides- space2 group p21212
|
|
|
| 105 | c5gafi_ |
|
not modelled |
52.0 |
17 |
PDB header:ribosome
Chain: I: PDB Molecule:50s ribosomal protein l10;
PDBTitle: rnc in complex with srp
|
|
|
| 106 | d1g5ta_ |
|
not modelled |
49.9 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
|
|
| 107 | d1g64b_ |
|
not modelled |
49.5 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
|
|
| 108 | c2j289_ |
|
not modelled |
47.7 |
14 |
PDB header:ribosome
Chain: 9: PDB Molecule:signal recognition particle 54;
PDBTitle: model of e. coli srp bound to 70s rncs
|
|
|
| 109 | c1vmaA_ |
|
not modelled |
47.5 |
24 |
PDB header:protein transport
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
|
|
|
| 110 | d1rl1a_ |
|
not modelled |
45.8 |
10 |
Fold:HSP20-like chaperones
Superfamily:HSP20-like chaperones
Family:GS domain |
|
|
| 111 | c3dmdA_ |
|
not modelled |
41.7 |
25 |
PDB header:transport protein
Chain: A: PDB Molecule:signal recognition particle receptor;
PDBTitle: structures and conformations in solution of the signal recognition2 particle receptor from the archaeon pyrococcus furiosus
|
|
|
| 112 | c3c0kB_ |
|
not modelled |
34.2 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:upf0064 protein yccw;
PDBTitle: crystal structure of a ribosomal rna methyltranferase
|
|
|
| 113 | c1wb1C_ |
|
not modelled |
32.4 |
12 |
PDB header:protein synthesis
Chain: C: PDB Molecule:translation elongation factor selb;
PDBTitle: crystal structure of translation elongation factor selb2 from methanococcus maripaludis in complex with gdp
|
|
|
| 114 | c6cy1B_ |
|
not modelled |
31.7 |
17 |
PDB header:signaling protein
Chain: B: PDB Molecule:signal recognition particle receptor ftsy;
PDBTitle: crystal structure of signal recognition particle receptor ftsy from2 elizabethkingia anophelis
|
|
|
| 115 | c2jkiS_ |
|
not modelled |
31.2 |
8 |
PDB header:chaperone
Chain: S: PDB Molecule:sgt1-like protein;
PDBTitle: complex of hsp90 n-terminal and sgt1 cs domain
|
|
|
| 116 | d1j8yf2 |
|
not modelled |
28.7 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 117 | c2as0A_ |
|
not modelled |
28.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical protein ph1915;
PDBTitle: crystal structure of ph1915 (apc 5817): a hypothetical rna2 methyltransferase
|
|
|
| 118 | c2cmgA_ |
|
not modelled |
26.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:spermidine synthase;
PDBTitle: crystal structure of spermidine synthase from helicobacter2 pylori
|
|
|
| 119 | d1lhpa_ |
|
not modelled |
25.5 |
21 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:PfkB-like kinase |
|
|
| 120 | d1ju2a1 |
|
not modelled |
25.4 |
16 |
Fold:FAD/NAD(P)-binding domain
Superfamily:FAD/NAD(P)-binding domain
Family:FAD-linked reductases, N-terminal domain |
|
|