| 1 | c2bpqB_
|
|
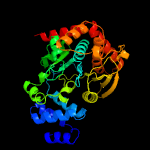 |
100.0 |
100 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase (trpd) from2 mycobacterium tuberculosis (apo structure)
|
|
|
|
| 2 | c1v8gB_
|
|
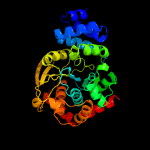 |
100.0 |
41 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase2 (trpd) from thermus thermophilus hb8
|
|
|
|
| 3 | c1khdD_
|
|
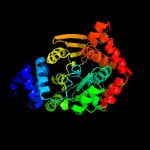 |
100.0 |
31 |
PDB header:transferase
Chain: D: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure analysis of the anthranilate2 phosphoribosyltransferase from erwinia carotovora at 1.9 resolution3 (current name, pectobacterium carotovorum)
|
|
|
|
| 4 | c4gtnA_
|
|
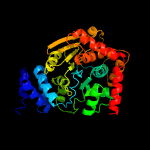 |
100.0 |
34 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: structure of anthranilate phosphoribosyl transferase from2 acinetobacter baylyi
|
|
|
|
| 5 | c5nofB_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyltransferase from thermococcus kodakaraensis
|
|
|
|
| 6 | c1o17A_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: anthranilate phosphoribosyl-transferase (trpd)
|
|
|
|
| 7 | c1vquB_
|
|
 |
100.0 |
38 |
PDB header:transferase
Chain: B: PDB Molecule:anthranilate phosphoribosyltransferase 2;
PDBTitle: crystal structure of anthranilate phosphoribosyltransferase 22 (17130499) from nostoc sp. at 1.85 a resolution
|
|
|
|
| 8 | c4hkmA_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:anthranilate phosphoribosyltransferase;
PDBTitle: crystal structure of an anthranilate phosphoribosyltransferase (target2 id nysgrc-016600) from xanthomonas campestris
|
|
|
|
| 9 | c4muoB_
|
|
 |
100.0 |
20 |
PDB header:dna binding protein
Chain: B: PDB Molecule:uncharacterized protein ybib;
PDBTitle: the trpd2 enzyme from e.coli: ybib
|
|
|
|
| 10 | c3h5qA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
|
|
|
| 11 | c4ga5H_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: H: PDB Molecule:putative thymidine phosphorylase;
PDBTitle: crystal structure of amp phosphorylase c-terminal deletion mutant in2 the apo-form
|
|
|
|
| 12 | c1brwB_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: B: PDB Molecule:protein (pyrimidine nucleoside phosphorylase);
PDBTitle: the crystal structure of pyrimidine nucleoside2 phosphorylase in a closed conformation
|
|
|
|
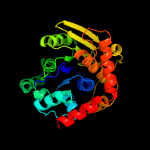
| 13 | d2elca2
|
|
 |
100.0 |
46 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
|
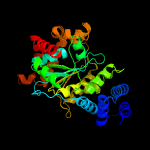
| 14 | c2j0fC_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: C: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural basis for non-competitive product inhibition in2 human thymidine phosphorylase: implication for drug design
|
|
|
|
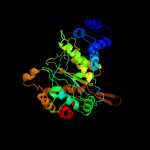
| 15 | c2dsjA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside (thymidine) phosphorylase;
PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
|
|
|
|
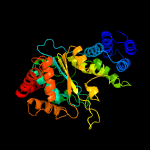
| 16 | c1otpA_
|
|
 |
100.0 |
18 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
|
|
|
| 17 | d1o17a2
|
|
 |
100.0 |
29 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
|
| 18 | d1khda2
|
|
 |
100.0 |
33 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
|
| 19 | d1uoua2
|
|
 |
100.0 |
22 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
|
| 20 | d1brwa2
|
|
 |
100.0 |
20 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
|
| 21 | d2tpta2 |
|
not modelled |
100.0 |
20 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 22 | d1khda1 |
|
not modelled |
99.7 |
23 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 23 | d1brwa1 |
|
not modelled |
99.7 |
19 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 24 | d2tpta1 |
|
not modelled |
99.7 |
20 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 25 | d1o17a1 |
|
not modelled |
99.7 |
24 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 26 | d1uoua1 |
|
not modelled |
99.7 |
18 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 27 | d1v8ga1 |
|
not modelled |
99.7 |
22 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 28 | d2elca1 |
|
not modelled |
99.3 |
20 |
Fold:Methionine synthase domain-like
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain |
|
|
| 29 | c4gijC_ |
|
not modelled |
85.3 |
22 |
PDB header:hydrolase
Chain: C: PDB Molecule:pseudouridine-5'-phosphate glycosidase;
PDBTitle: crystal structure of pseudouridine monophosphate glycosidase complexed2 with sulfate
|
|
|
| 30 | c1qzwC_ |
|
not modelled |
85.0 |
14 |
PDB header:signaling protein/rna
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the complete core of archaeal srp and2 implications for inter-domain communication
|
|
|
| 31 | d1vkma_ |
|
not modelled |
84.7 |
18 |
Fold:Indigoidine synthase A-like
Superfamily:Indigoidine synthase A-like
Family:Indigoidine synthase A-like |
|
|
| 32 | c1vmaA_ |
|
not modelled |
80.9 |
15 |
PDB header:protein transport
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: crystal structure of cell division protein ftsy (tm0570) from2 thermotoga maritima at 1.60 a resolution
|
|
|
| 33 | c4ex8A_ |
|
not modelled |
80.6 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:alna;
PDBTitle: crystal structure of the prealnumycin c-glycosynthase alna
|
|
|
| 34 | c5zmfA_ |
|
not modelled |
78.6 |
17 |
PDB header:hydrolase/transport protein
Chain: A: PDB Molecule:atpase arsa1;
PDBTitle: amppnp complex of c. reinhardtii arsa1
|
|
|
| 35 | d1p88a_ |
|
not modelled |
77.5 |
17 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
| 36 | c2v3cC_ |
|
not modelled |
74.1 |
12 |
PDB header:signaling protein
Chain: C: PDB Molecule:signal recognition 54 kda protein;
PDBTitle: crystal structure of the srp54-srp19-7s.s srp rna complex2 of m. jannaschii
|
|
|
| 37 | c2qguA_ |
|
not modelled |
71.5 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:probable signal peptide protein;
PDBTitle: three-dimensional structure of the phospholipid-binding protein from2 ralstonia solanacearum q8xv73_ralsq in complex with a phospholipid at3 the resolution 1.53 a. northeast structural genomics consortium4 target rsr89
|
|
|
| 38 | c2yhsA_ |
|
not modelled |
70.3 |
13 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of the e. coli srp receptor ftsy
|
|
|
| 39 | c3zh3A_ |
|
not modelled |
67.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: crystal structure of s. pneumoniae d39 native mura1
|
|
|
| 40 | c2kvcA_ |
|
not modelled |
65.7 |
15 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
|
|
|
| 41 | c3zq6D_ |
|
not modelled |
64.6 |
23 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative arsenical pump-driving atpase;
PDBTitle: adp-alf4 complex of m. therm. trc40
|
|
|
| 42 | c2m0nA_ |
|
not modelled |
62.8 |
21 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of a duf3349 annotated protein from mycobacterium2 abscessus, mab_3403c. seattle structural genomics center for3 infectious disease target myaba.17112.a.a2
|
|
|
| 43 | c5bufA_ |
|
not modelled |
62.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: 2.37 angstrom structure of epsp synthase from acinetobacter baumannii
|
|
|
| 44 | d1ihua2 |
|
not modelled |
62.0 |
25 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 45 | c6bs3A_ |
|
not modelled |
61.4 |
29 |
PDB header:unknown function
Chain: A: PDB Molecule:putative atpase rv3679;
PDBTitle: crystal structure of adp-bound bacterial get3-like a and b in2 mycobacterium tuberculosis
|
|
|
| 46 | c6bs5B_ |
|
not modelled |
61.1 |
34 |
PDB header:unknown function
Chain: B: PDB Molecule:anion transporter;
PDBTitle: crystal structure of amp-pnp-bound bacterial get3-like a and b in2 mycobacterium tuberculosis
|
|
|
| 47 | c3kc2A_ |
|
not modelled |
59.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ykr070w;
PDBTitle: crystal structure of mitochondrial had-like phosphatase from2 saccharomyces cerevisiae
|
|
|
| 48 | d2afhe1 |
|
not modelled |
57.9 |
25 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 49 | c3endA_ |
|
not modelled |
57.7 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:light-independent protochlorophyllide reductase
PDBTitle: crystal structure of the l protein of rhodobacter2 sphaeroides light-independent protochlorophyllide3 reductase (bchl) with mgadp bound: a homologue of the4 nitrogenase fe protein
|
|
|
| 50 | c2lkyA_ |
|
not modelled |
57.0 |
9 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
|
|
|
| 51 | c5dcfA_ |
|
not modelled |
56.9 |
13 |
PDB header:recombination
Chain: A: PDB Molecule:tyrosine recombinase xerd,dna translocase ftsk;
PDBTitle: c-terminal domain of xerd recombinase in complex with gamma domain of2 ftsk
|
|
|
| 52 | c5uwaB_ |
|
not modelled |
56.4 |
11 |
PDB header:transport protein
Chain: B: PDB Molecule:probable phospholipid-binding protein mlac;
PDBTitle: structure of e. coli phospholipid binding protein mlac
|
|
|
| 53 | c5lwcA_ |
|
not modelled |
55.4 |
21 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:bacteriocin bacsp222;
PDBTitle: nmr solution structure of bacteriocin bacsp222 from staphylococcus2 pseudintermedius 222
|
|
|
| 54 | c4rtbA_ |
|
not modelled |
55.0 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
| 55 | c2pqdA_ |
|
not modelled |
54.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: a100g cp4 epsps liganded with (r)-difluoromethyl tetrahedral reaction2 intermediate analog
|
|
|
| 56 | c5ujsB_ |
|
not modelled |
53.5 |
14 |
PDB header:hydrolase,oxidoreductase
Chain: B: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: 2.45 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase from campylobacter jejuni.
|
|
|
| 57 | c3ug7D_ |
|
not modelled |
53.4 |
31 |
PDB header:hydrolase
Chain: D: PDB Molecule:arsenical pump-driving atpase;
PDBTitle: crystal structure of get3 from methanocaldococcus jannaschii
|
|
|
| 58 | d1cp2a_ |
|
not modelled |
53.4 |
26 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 59 | c3ibgF_ |
|
not modelled |
51.3 |
34 |
PDB header:hydrolase
Chain: F: PDB Molecule:atpase, subunit of the get complex;
PDBTitle: crystal structure of aspergillus fumigatus get3 with bound2 adp
|
|
|
| 60 | c5wi5C_ |
|
not modelled |
51.1 |
23 |
PDB header:transferase
Chain: C: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.0 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase from streptococcus pneumoniae in complex3 with uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2r)-4 2-(phosphonooxy)propanoic acid and magnesium.
|
|
|
| 61 | d1rf6a_ |
|
not modelled |
49.6 |
23 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
| 62 | c1ii0A_ |
|
not modelled |
47.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:arsenical pump-driving atpase;
PDBTitle: crystal structure of the escherichia coli arsenite-translocating2 atpase
|
|
|
| 63 | d1a0pa2 |
|
not modelled |
47.6 |
13 |
Fold:DNA breaking-rejoining enzymes
Superfamily:DNA breaking-rejoining enzymes
Family:Lambda integrase-like, catalytic core |
|
|
| 64 | c2qy9A_ |
|
not modelled |
47.5 |
13 |
PDB header:protein transport
Chain: A: PDB Molecule:cell division protein ftsy;
PDBTitle: structure of the ng+1 construct of the e. coli srp receptor2 ftsy
|
|
|
| 65 | d1g6sa_ |
|
not modelled |
46.6 |
17 |
Fold:IF3-like
Superfamily:EPT/RTPC-like
Family:Enolpyruvate transferase, EPT |
|
|
| 66 | c2iy3A_ |
|
not modelled |
45.9 |
14 |
PDB header:rna-binding
Chain: A: PDB Molecule:signal recognition particle protein,signal recognition
PDBTitle: structure of the e. coli signal regognition particle
|
|
|
| 67 | c1r71B_ |
|
not modelled |
45.9 |
15 |
PDB header:transcription/dna
Chain: B: PDB Molecule:transcriptional repressor protein korb;
PDBTitle: crystal structure of the dna binding domain of korb in complex with2 the operator dna
|
|
|
| 68 | c2wooC_ |
|
not modelled |
45.2 |
31 |
PDB header:hydrolase
Chain: C: PDB Molecule:atpase get3;
PDBTitle: nucleotide-free form of s. pombe get3
|
|
|
| 69 | c5xwbB_ |
|
not modelled |
44.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase;
PDBTitle: crystal structure of 5-enolpyruvulshikimate-3-phosphate synthase from2 a psychrophilic bacterium, colwellia psychrerythraea
|
|
|
| 70 | c5gafi_ |
|
not modelled |
44.2 |
15 |
PDB header:ribosome
Chain: I: PDB Molecule:50s ribosomal protein l10;
PDBTitle: rnc in complex with srp
|
|
|
| 71 | d1j9ia_ |
|
not modelled |
43.8 |
19 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:Terminase gpNU1 subunit domain |
|
|
| 72 | c3rmtB_ |
|
not modelled |
43.1 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:3-phosphoshikimate 1-carboxyvinyltransferase 1;
PDBTitle: crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate2 synthase from bacillus halodurans c-125
|
|
|
| 73 | c3igfB_ |
|
not modelled |
43.0 |
23 |
PDB header:atp binding protein
Chain: B: PDB Molecule:all4481 protein;
PDBTitle: crystal structure of the all4481 protein from nostoc sp. pcc 7120,2 northeast structural genomics consortium target nsr300
|
|
|
| 74 | c4wcxC_ |
|
not modelled |
43.0 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
| 75 | c2j37W_ |
|
not modelled |
41.3 |
9 |
PDB header:ribosome
Chain: W: PDB Molecule:signal recognition particle 54 kda protein (srp54);
PDBTitle: model of mammalian srp bound to 80s rncs
|
|
|
| 76 | c4ru8C_ |
|
not modelled |
41.1 |
32 |
PDB header:hydrolase
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: structure of pnob8 para with amppnp
|
|
|
| 77 | c3eegB_ |
|
not modelled |
39.6 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of a 2-isopropylmalate synthase from2 cytophaga hutchinsonii
|
|
|
| 78 | c2mqkA_ |
|
not modelled |
39.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent target dna activator b;
PDBTitle: solution structure of n terminal domain of the mub aaa+ atpase
|
|
|
| 79 | d1w4ta1 |
|
not modelled |
38.7 |
18 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Arylamine N-acetyltransferase |
|
|
| 80 | d1byia_ |
|
not modelled |
38.0 |
30 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 81 | c3vx3A_ |
|
not modelled |
37.7 |
41 |
PDB header:adp binding protein
Chain: A: PDB Molecule:atpase involved in chromosome partitioning, para/mind
PDBTitle: crystal structure of [nife] hydrogenase maturation protein hypb from2 thermococcus kodakarensis kod1
|
|
|
| 82 | d1r71a_ |
|
not modelled |
37.0 |
16 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:KorB DNA-binding domain-like
Family:KorB DNA-binding domain-like |
|
|
| 83 | c4gxwA_ |
|
not modelled |
35.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosine deaminase;
PDBTitle: crystal structure of a cog1816 amidohydrolase (target efi-505188) from2 burkhoderia ambifaria, with bound zn
|
|
|
| 84 | c3of5A_ |
|
not modelled |
34.5 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:dethiobiotin synthetase;
PDBTitle: crystal structure of a dethiobiotin synthetase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 85 | d1b8za_ |
|
not modelled |
33.7 |
10 |
Fold:IHF-like DNA-binding proteins
Superfamily:IHF-like DNA-binding proteins
Family:Prokaryotic DNA-bending protein |
|
|
| 86 | c2j289_ |
|
not modelled |
33.5 |
14 |
PDB header:ribosome
Chain: 9: PDB Molecule:signal recognition particle 54;
PDBTitle: model of e. coli srp bound to 70s rncs
|
|
|
| 87 | c3bg3B_ |
|
not modelled |
32.8 |
17 |
PDB header:ligase
Chain: B: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
| 88 | c3katA_ |
|
not modelled |
32.7 |
20 |
PDB header:apoptosis
Chain: A: PDB Molecule:nacht, lrr and pyd domains-containing protein 1;
PDBTitle: crystal structure of the card domain of the human nlrp1 protein,2 northeast structural genomics consortium target hr3486e
|
|
|
| 89 | c5lvtC_ |
|
not modelled |
32.2 |
6 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna-binding protein hu;
PDBTitle: structure of hu protein from lactococcus lactis
|
|
|
| 90 | d1mula_ |
|
not modelled |
32.1 |
17 |
Fold:IHF-like DNA-binding proteins
Superfamily:IHF-like DNA-binding proteins
Family:Prokaryotic DNA-bending protein |
|
|
| 91 | c1nvmG_ |
|
not modelled |
31.7 |
15 |
PDB header:lyase/oxidoreductase
Chain: G: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of a bifunctional aldolase-dehydrogenase :2 sequestering a reactive and volatile intermediate
|
|
|
| 92 | c2yvqA_ |
|
not modelled |
31.1 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
|
|
| 93 | c3kjgB_ |
|
not modelled |
30.9 |
22 |
PDB header:hydrolase, metal binding protein
Chain: B: PDB Molecule:co dehydrogenase/acetyl-coa synthase complex, accessory
PDBTitle: adp-bound state of cooc1
|
|
|
| 94 | c2ozeA_ |
|
not modelled |
30.4 |
36 |
PDB header:dna binding protein
Chain: A: PDB Molecule:orf delta';
PDBTitle: the crystal structure of delta protein of psm19035 from2 streptoccocus pyogenes
|
|
|
| 95 | c3r38A_ |
|
not modelled |
29.8 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;
PDBTitle: 2.23 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase (mura) from listeria monocytogenes egd-e
|
|
|
| 96 | c3bg3A_ |
|
not modelled |
29.6 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:pyruvate carboxylase, mitochondrial;
PDBTitle: crystal structure of human pyruvate carboxylase (missing the biotin2 carboxylase domain at the n-terminus)
|
|
|
| 97 | c3ol4B_ |
|
not modelled |
29.4 |
25 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
|
|
|
| 98 | c2ndpA_ |
|
not modelled |
28.9 |
10 |
PDB header:dna binding protein
Chain: A: PDB Molecule:histone-like dna-binding superfamily protein;
PDBTitle: structure of dna-binding hu protein from micoplasma mycoplasma2 gallisepticum
|
|
|
| 99 | c6drpB_ |
|
not modelled |
28.8 |
11 |
PDB header:immune system
Chain: B: PDB Molecule:nlr family card domain-containing protein 4;
PDBTitle: cryo-em structures of asc and nlrc4 card filaments reveal a unified2 mechanism of nucleation and activation of caspase-1
|
|
|
| 100 | c5hy7D_ |
|
not modelled |
28.6 |
15 |
PDB header:protein binding
Chain: D: PDB Molecule:ysf3;
PDBTitle: sf3b10-sf3b130 from chaetomium thermophilum
|
|
|
| 101 | c5ks8D_ |
|
not modelled |
28.4 |
20 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 102 | d1ihua1 |
|
not modelled |
28.3 |
24 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 103 | c4ak9A_ |
|
not modelled |
28.3 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:cpftsy;
PDBTitle: structure of chloroplast ftsy from physcomitrella patens
|
|
|
| 104 | c5bq2C_ |
|
not modelled |
28.1 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:udp-n-acetylglucosamine 1-carboxyvinyltransferase;
PDBTitle: crystal structure of udp-n-acetylglucosamine 1-carboxyvinyltransferase2 (udp-n-acetylglucosamine enolpyruvyl transferase, ept) from3 pseudomonas aeruginosa
|
|
|
| 105 | c1rr2A_ |
|
not modelled |
28.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
|
|
| 106 | c2dg2D_ |
|
not modelled |
27.4 |
15 |
PDB header:protein binding
Chain: D: PDB Molecule:apolipoprotein a-i binding protein;
PDBTitle: crystal structure of mouse apolipoprotein a-i binding protein
|
|
|
| 107 | c3rhiB_ |
|
not modelled |
27.4 |
7 |
PDB header:dna binding protein
Chain: B: PDB Molecule:dna-binding protein hu;
PDBTitle: dna-binding protein hu from bacillus anthracis
|
|
|
| 108 | c2o8bA_ |
|
not modelled |
27.4 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:dna mismatch repair protein msh2;
PDBTitle: human mutsalpha (msh2/msh6) bound to adp and a g t mispair
|
|
|
| 109 | c3ewbX_ |
|
not modelled |
27.2 |
14 |
PDB header:transferase
Chain: X: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of n-terminal domain of putative 2-isopropylmalate2 synthase from listeria monocytogenes
|
|
|
| 110 | c1tuuA_ |
|
not modelled |
27.2 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:acetate kinase;
PDBTitle: acetate kinase crystallized with atpgs
|
|
|
| 111 | c2xj9B_ |
|
not modelled |
27.1 |
24 |
PDB header:replication
Chain: B: PDB Molecule:mipz;
PDBTitle: dimer structure of the bacterial cell division regulator mipz
|
|
|
| 112 | c3stgA_ |
|
not modelled |
26.3 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:2-dehydro-3-deoxyphosphooctonate aldolase;
PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
|
|
|
| 113 | c2ph1A_ |
|
not modelled |
26.0 |
29 |
PDB header:ligand binding protein
Chain: A: PDB Molecule:nucleotide-binding protein;
PDBTitle: crystal structure of nucleotide-binding protein af2382 from2 archaeoglobus fulgidus, northeast structural genomics target gr165
|
|
|
| 114 | c1hyqA_ |
|
not modelled |
25.8 |
28 |
PDB header:cell cycle
Chain: A: PDB Molecule:cell division inhibitor (mind-1);
PDBTitle: mind bacterial cell division regulator from a. fulgidus
|
|
|
| 115 | d1hyqa_ |
|
not modelled |
25.8 |
28 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 116 | c6iucC_ |
|
not modelled |
25.6 |
24 |
PDB header:dna binding protein/dna
Chain: C: PDB Molecule:spooj regulator (soj);
PDBTitle: structure of helicobacter pylori soj-atp complex bound to dna
|
|
|
| 117 | d1tuea_ |
|
not modelled |
25.5 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
| 118 | c4fczB_ |
|
not modelled |
25.5 |
6 |
PDB header:transport protein
Chain: B: PDB Molecule:toluene-tolerance protein;
PDBTitle: crystal structure of toluene-tolerance protein from pseudomonas putida2 (strain kt2440), northeast structural genomics consortium (nesg)3 target ppr99
|
|
|
| 119 | c5ekaA_ |
|
not modelled |
25.5 |
7 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna-binding protein hu;
PDBTitle: hu dna-binding protein from thermus thermophilus
|
|
|
| 120 | c5x9wA_ |
|
not modelled |
25.2 |
17 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna mismatch repair protein muts;
PDBTitle: mismatch repair protein
|
|
|




























































































































































































































































