| 1 |
|
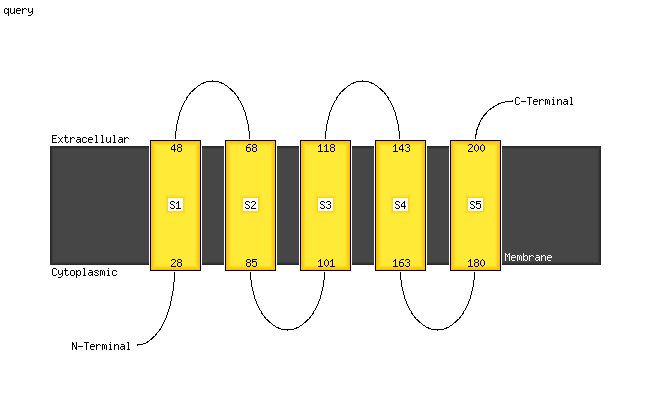
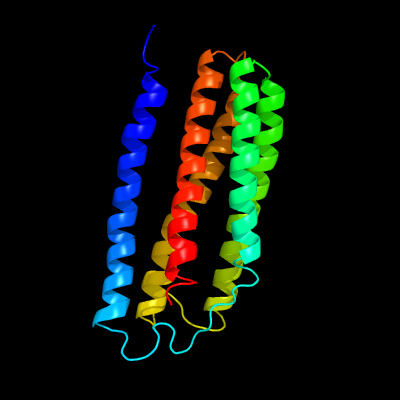
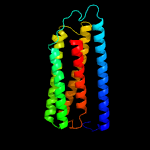
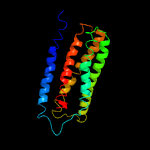
PDB 6hwh chain W
Region: 20 - 203
Aligned: 184
Modelled: 184
Confidence: 100.0%
Identity: 87%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase subunit 3;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
Phyre2
| 2 |
|
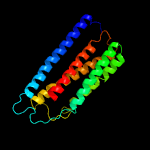
PDB 2yev chain A
Region: 7 - 201
Aligned: 192
Modelled: 195
Confidence: 100.0%
Identity: 27%
PDB header:electron transport
Chain: A: PDB Molecule:cytochrome c oxidase polypeptide i+iii;
PDBTitle: structure of caa3-type cytochrome oxidase
Phyre2
| 3 |
|
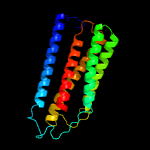
PDB 1v54 chain C
Region: 11 - 201
Aligned: 188
Modelled: 191
Confidence: 100.0%
Identity: 33%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 4 |
|
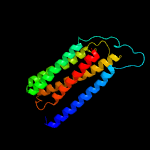
PDB 1qle chain C
Region: 13 - 201
Aligned: 186
Modelled: 189
Confidence: 100.0%
Identity: 31%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 5 |
|
PDB 6hu9 chain O
Region: 11 - 201
Aligned: 188
Modelled: 191
Confidence: 100.0%
Identity: 28%
PDB header:oxidoreductase/electron transport
Chain: O: PDB Molecule:cytochrome c1, heme protein, mitochondrial;
PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
Phyre2
| 6 |
|
PDB 1m56 chain C
Region: 13 - 201
Aligned: 186
Modelled: 189
Confidence: 100.0%
Identity: 30%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 7 |
|
PDB 1fft chain C
Region: 19 - 202
Aligned: 183
Modelled: 184
Confidence: 100.0%
Identity: 30%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 8 |
|
PDB 5tc1 chain M
Region: 77 - 103
Aligned: 27
Modelled: 27
Confidence: 10.8%
Identity: 22%
PDB header:viral protein/rna
Chain: M: PDB Molecule:maturation protein;
PDBTitle: in situ structures of the genome and genome-delivery apparatus in2 ssrna bacteriophage ms2
Phyre2
| 9 |
|
PDB 1jb0 chain I
Region: 102 - 117
Aligned: 16
Modelled: 16
Confidence: 10.1%
Identity: 38%
Fold: Single transmembrane helix
Superfamily: Subunit VIII of photosystem I reaction centre, PsaI
Family: Subunit VIII of photosystem I reaction centre, PsaI
Phyre2
| 10 |
|
PDB 4fe1 chain I
Region: 102 - 117
Aligned: 16
Modelled: 16
Confidence: 10.1%
Identity: 38%
PDB header:photosynthesis
Chain: I: PDB Molecule:photosystem i reaction center subunit viii;
PDBTitle: improving the accuracy of macromolecular structure refinement at 7 a2 resolution
Phyre2
| 11 |
|
PDB 4dja chain A
Region: 181 - 189
Aligned: 9
Modelled: 9
Confidence: 9.0%
Identity: 33%
PDB header:lyase
Chain: A: PDB Molecule:photolyase;
PDBTitle: crystal structure of a prokaryotic (6-4) photolyase phrb from2 agrobacterium tumefaciens with an fe-s cluster and a 6,7-dimethyl-8-3 ribityllumazine antenna chromophore at 1.45a resolution
Phyre2
| 12 |
|
PDB 4cso chain D
Region: 182 - 190
Aligned: 9
Modelled: 9
Confidence: 8.4%
Identity: 44%
PDB header:transcription
Chain: D: PDB Molecule:orfy protein, transcription factor;
PDBTitle: the structure of orfy from thermoproteus tenax
Phyre2
| 13 |
|
PDB 6hq8 chain B
Region: 180 - 188
Aligned: 9
Modelled: 9
Confidence: 7.9%
Identity: 56%
PDB header:hydrolase
Chain: B: PDB Molecule:beta-1,3-oligosaccharide phosphorylase;
PDBTitle: bacterial beta-1,3-oligosaccharide phosphorylase from gh149 with2 laminarihexaose bound at a surface site
Phyre2
| 14 |
|
PDB 5mnt chain D
Region: 77 - 103
Aligned: 27
Modelled: 27
Confidence: 6.6%
Identity: 33%
PDB header:rna binding protein
Chain: D: PDB Molecule:a2 maturation protein;
PDBTitle: bacteriophage qbeta maturation protein
Phyre2