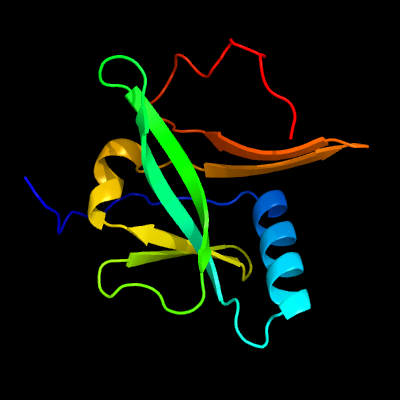
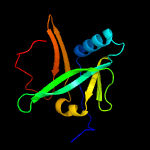
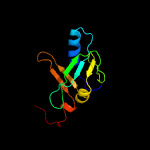
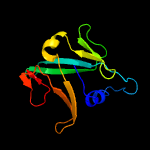
1 c2d2aA_
100.0
32
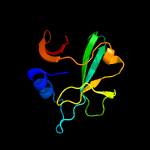
PDB header: metal transportChain: A: PDB Molecule: sufa protein;PDBTitle: crystal structure of escherichia coli sufa involved in biosynthesis of2 iron-sulfur clusters
2 c2apnA_
100.0
43
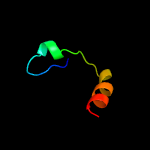
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein hi1723;PDBTitle: hi1723 solution structure
3 c1x0gA_
100.0
32
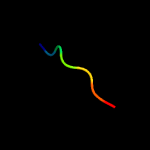
PDB header: metal binding proteinChain: A: PDB Molecule: isca;PDBTitle: crystal structure of isca with the [2fe-2s] cluster
4 c2k4zA_
100.0
23
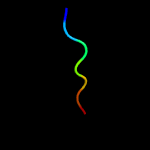
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: dsrr;PDBTitle: solution nmr structure of allochromatium vinosum dsrr:2 northeast structural genomics consortium target op5
5 d1s98a_
100.0
33
Fold: HesB-like domainSuperfamily: HesB-like domainFamily: HesB-like domain
6 d1nwba_
100.0
37
Fold: HesB-like domainSuperfamily: HesB-like domainFamily: HesB-like domain
7 d2p2ea1
98.1
16
Fold: HesB-like domainSuperfamily: HesB-like domainFamily: HesB-like domain
8 c2qgoA_
97.9
14
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative fe-s biosynthesis protein;PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
9 d1sp2a_
17.4
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
10 c2x5hD_
16.2
18
PDB header: viral proteinChain: D: PDB Molecule: orf 131;PDBTitle: crystal structure of the orf131 l26m l51m double mutant2 from sulfolobus islandicus rudivirus 1
11 d1zfda_
16.0
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
12 d1a1ia1
13.9
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
13 c2ab3A_
13.8
83
PDB header: rna binding proteinChain: A: PDB Molecule: znf29;PDBTitle: solution structures and characterization of hiv rre iib rna2 targeting zinc finger proteins
14 c6bzsA_
13.7
13
PDB header: transport proteinChain: A: PDB Molecule: multidrug resistance-associated protein 6;PDBTitle: human abcc6 nbd1 in apo state
15 d1aaya1
13.3
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
16 d1a1ga1
12.9
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
17 d1ubdc3
12.8
43
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
18 c1gxsC_
12.6
19
PDB header: lyaseChain: C: PDB Molecule: p-(s)-hydroxymandelonitrile lyase chain a;PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
19 d1u86a1
12.3
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
20 d1a1ha1
12.2
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
21 c4imhA_
not modelled
12.1
20
PDB header: metal transport, transport proteinChain: A: PDB Molecule: hemin degrading factor;PDBTitle: crystal structure of cytoplasmic heme binding protein, phus, from2 pseudomonas aeruginosa
22 d2hq2a1
not modelled
12.0
23
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like
23 d1f2ig1
not modelled
11.6
38
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
24 d1tf3a2
not modelled
11.1
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
25 c2kvfA_
not modelled
11.1
30
PDB header: transcriptionChain: A: PDB Molecule: zinc finger and btb domain-containing protein 32;PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
26 d2glia3
not modelled
10.8
21
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
27 c4z4pA_
not modelled
10.4
32
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase 2d;PDBTitle: structure of the mll4 set domain
28 d2j0pa1
not modelled
10.1
20
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like
29 c4ox3A_
not modelled
10.0
18
PDB header: hydrolaseChain: A: PDB Molecule: putative carboxypeptidase yodj;PDBTitle: structure of the ldcb ld-carboxypeptidase reveals the molecular basis2 of peptidoglycan recognition
30 d2cy5a1
not modelled
9.9
22
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Phosphotyrosine-binding domain (PTB)
31 c3tsjA_
not modelled
9.8
18
PDB header: allergen, oxidoreductaseChain: A: PDB Molecule: pollen allergen phl p 4;PDBTitle: crystal structure of phl p 4, a grass pollen allergen with glucose2 dehydrogenase activity
32 d2yt9a1
not modelled
9.6
43
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
33 c6n2gC_
not modelled
9.1
10
PDB header: chaperoneChain: C: PDB Molecule: nucleosome assembly protein;PDBTitle: crystal structure of caenorhabditis elegans nap1
34 d1ncsa_
not modelled
8.7
29
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
35 d2dlka2
not modelled
8.5
86
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
36 c3qkgA_
not modelled
8.4
15
PDB header: immune systemChain: A: PDB Molecule: protein ambp;PDBTitle: crystal structure of alpha-1-microglobulin at 2.3 a resolution
37 c2kdxA_
not modelled
8.4
18
PDB header: metal-binding proteinChain: A: PDB Molecule: hydrogenase/urease nickel incorporation proteinPDBTitle: solution structure of hypa protein
38 d1x6ea2
not modelled
8.1
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
39 c5b3hC_
not modelled
8.0
50
PDB header: transcriptionChain: C: PDB Molecule: zinc finger protein jackdaw;PDBTitle: the crystal structure of the jackdaw/idd10 bound to the heterodimeric2 shr-scr complex
40 c3pm7A_
not modelled
8.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of ef_3132 protein from enterococcus faecalis at the2 resolution 2a, northeast structural genomics consortium target efr184
41 d2a2ua_
not modelled
8.0
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like
42 c2nq2C_
not modelled
7.8
11
PDB header: metal transportChain: C: PDB Molecule: hypothetical abc transporter atp-binding proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
43 c5zet2_
not modelled
7.7
71
PDB header: ribosomeChain: 2: PDB Molecule: 50s ribosomal protein l31;PDBTitle: m. smegmatis p/p state 50s ribosomal subunit
44 c1bcrA_
not modelled
7.6
21
PDB header: hydrolase/hydrolase inhibitorChain: A: PDB Molecule: serine carboxypeptidase ii;PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
45 c4az3A_
not modelled
7.5
17
PDB header: hydrolaseChain: A: PDB Molecule: lysosomal protective protein 32 kda chain;PDBTitle: crystal structure of cathepsin a, complexed with 15a
46 c2wh44_
not modelled
7.5
60
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome
47 c5gplA_
not modelled
7.3
19
PDB header: chaperoneChain: A: PDB Molecule: putative nucleosome assembly protein c36b7.08c;PDBTitle: crystal structure of ccp1
48 d1vsra_
not modelled
7.2
10
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Very short patch repair (VSR) endonuclease
49 c2l3aA_
not modelled
7.1
7
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of homodimer protein sp_0782 (7-79) from2 streptococcus pneumoniae northeast structural genomics consortium3 target spr104 .
50 c1znmA_
not modelled
7.1
43
PDB header: zinc fingerChain: A: PDB Molecule: yy1;PDBTitle: a zinc finger with an artificial beta-turn, original sequence taken2 from the third zinc finger domain of the human transcriptional3 repressor protein yy1 (ying and yang 1, a delta transcription4 factor), nmr, 34 structures
51 c3a44D_
not modelled
7.0
17
PDB header: metal binding proteinChain: D: PDB Molecule: hydrogenase nickel incorporation protein hypa;PDBTitle: crystal structure of hypa in the dimeric form
52 c3c3jA_
not modelled
6.8
25
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
53 c3f1f4_
not modelled
6.8
60
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: crystal structure of a translation termination complex formed with2 release factor rf2. this file contains the 50s subunit of one 70s3 ribosome. the entire crystal structure contains two 70s ribosomes as4 described in remark 400.
54 c5ghrA_
not modelled
6.8
23
PDB header: dna binding protein/replicationChain: A: PDB Molecule: ssdna-specific exonuclease;PDBTitle: dna replication protein
55 d1jzua_
not modelled
6.7
18
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like
56 c2yujA_
not modelled
6.7
20
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin fusion degradation 1-like;PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
57 c1va1A_
not modelled
6.7
29
PDB header: transcriptionChain: A: PDB Molecule: transcription factor sp1;PDBTitle: solution structure of transcription factor sp1 dna binding2 domain (zinc finger 1)
58 c4lrvL_
not modelled
6.6
21
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
59 c5o60g_
not modelled
6.6
71
PDB header: ribosomeChain: G: PDB Molecule: 50s ribosomal protein l6;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
60 c3h5jA_
not modelled
6.5
40
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: leud_1-168 small subunit of isopropylmalate isomerase (rv2987c) from2 mycobacterium tuberculosis
61 c6avjB_
not modelled
6.3
15
PDB header: metal binding proteinChain: B: PDB Molecule: cdgsh iron-sulfur domain-containing protein 3,PDBTitle: crystal structure of human mitochondrial inner neet protein (mint)2 /cisd3
62 c3a0rA_
not modelled
6.2
14
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
63 c3q3wB_
not modelled
6.2
40
PDB header: transferaseChain: B: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
64 c2hcuA_
not modelled
6.2
40
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus mutans
65 c1t2yA_
not modelled
6.0
36
PDB header: metal binding proteinChain: A: PDB Molecule: metallothionein;PDBTitle: nmr solution structure of the protein part of cu6-2 neurospora crassa mt
66 d2cota1
not modelled
6.0
50
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
67 d1r44a_
not modelled
5.9
12
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanX-like
68 d1znda1
not modelled
5.9
12
Fold: LipocalinsSuperfamily: LipocalinsFamily: Retinol binding protein-like
69 c3odpA_
not modelled
5.8
25
PDB header: isomeraseChain: A: PDB Molecule: putative tagatose-6-phosphate ketose/aldose isomerase;PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
70 d1gq2a2
not modelled
5.8
14
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Malic enzyme N-domain
71 c2j034_
not modelled
5.6
43
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l31;PDBTitle: structure of the thermus thermophilus 70s ribosome2 complexed with mrna, trna and paromomycin (part 4 of 4).3 this file contains the 50s subunit from molecule ii.
72 d2j0141
not modelled
5.6
43
Fold: L28p-likeSuperfamily: L28p-likeFamily: Ribosomal protein L31p
73 d2h85a2
not modelled
5.6
29
Fold: EndoU-likeSuperfamily: EndoU-likeFamily: Nsp15 C-terminal domain-like
74 d1vmha_
not modelled
5.5
14
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
75 d1wmia1
not modelled
5.5
20
Fold: RelE-likeSuperfamily: RelE-likeFamily: RelE-like
76 c6b3qa_
not modelled
5.5
75
PDB header: hydrolase/hormoneChain: A: PDB Molecule: insulin-degrading enzyme;PDBTitle: cryo-em structure of human insulin degrading enzyme in complex with2 insulin
77 c3hlzA_
not modelled
5.4
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein bt_1490;PDBTitle: crystal structure of bt_1490 (np_810393.1) from bacteroides2 thetaiotaomicron vpi-5482 at 1.50 a resolution
78 d1ivya_
not modelled
5.4
16
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Serine carboxypeptidase-like
79 c5hnmC_
not modelled
5.4
16
PDB header: hydrolaseChain: C: PDB Molecule: d-alanyl-d-alanine carboxypeptidase;PDBTitle: crystal structure of vancomycin resistance d,d-pentapeptidase vany2 e175a mutant from vanb-type resistance cassette in complex with3 zn(ii)
80 d2cota2
not modelled
5.3
67
Fold: beta-beta-alpha zinc fingersSuperfamily: beta-beta-alpha zinc fingersFamily: Classic zinc finger, C2H2
81 c4f7uP_
not modelled
5.3
11
PDB header: splicingChain: P: PDB Molecule: methylosome subunit picln;PDBTitle: the 6s snrnp assembly intermediate
82 c3vbaE_
not modelled
5.3
60
PDB header: lyaseChain: E: PDB Molecule: isopropylmalate/citramalate isomerase small subunit;PDBTitle: crystal structure of methanogen 3-isopropylmalate isomerase small2 subunit
83 d1jj7a_
not modelled
5.3
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
84 d2ayua1
not modelled
5.2
16
Fold: NAP-likeSuperfamily: NAP-likeFamily: NAP-like
85 c2ayuA_
not modelled
5.2
16
PDB header: chaperoneChain: A: PDB Molecule: nucleosome assembly protein;PDBTitle: the structure of nucleosome assembly protein suggests a mechanism for2 histone binding and shuttling
86 d2jn4a1
not modelled
5.2
12
Fold: NifT/FixU barrel-likeSuperfamily: NifT/FixU-likeFamily: NifT/FixU
87 c2jn4A_
not modelled
5.2
12
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein fixu, nift;PDBTitle: solution nmr structure of protein rp4601 from rhodopseudomonas2 palustris. northeast structural genomics consortium target rpt2;3 ontario center for structural proteomics target rp4601.
88 d1p4xa1
not modelled
5.2
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MarR-like transcriptional regulators
89 c5vbnB_
not modelled
5.1
19
PDB header: transferaseChain: B: PDB Molecule: dna polymerase epsilon catalytic subunit a;PDBTitle: crystal structure of human dna polymerase epsilon b-subunit in complex2 with c-terminal domain of catalytic subunit