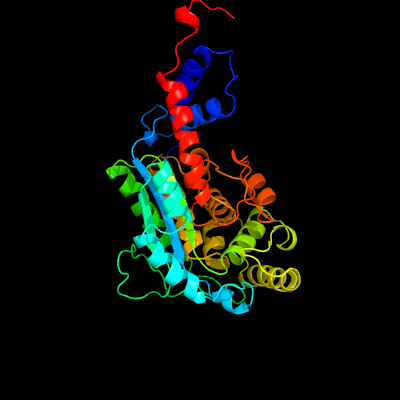
| 1 | d1l5oa_
|
|
 |
100.0 |
34 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
|
|
|
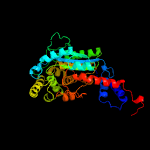
| 2 | d1j33a_
|
|
 |
100.0 |
50 |
Fold:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Superfamily:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)
Family:Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) |
|
|
|
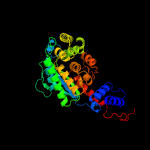
| 3 | c6b5fB_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: B: PDB Molecule:nicotinate-nucleotide--dimethylbenzimidazole
PDBTitle: crystal structure of nicotinate mononucleotide-5,6-2 dimethylbenzimidazole phosphoribosyltransferase cobt from yersinia3 enterocolitica
|
|
|
|
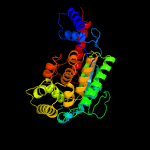
| 4 | c4hdnA_
|
|
 |
100.0 |
35 |
PDB header:transferase
Chain: A: PDB Molecule:arsa;
PDBTitle: crystal structure of arsab in the substrate-free state.
|
|
|
|
| 5 | c4hdnB_
|
|
 |
100.0 |
34 |
PDB header:transferase
Chain: B: PDB Molecule:arsb;
PDBTitle: crystal structure of arsab in the substrate-free state.
|
|
|
|
| 6 | c3l0zC_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:putative nicotinate-nucleotide-dimethylbenzimidazole
PDBTitle: crystal structure of a putative nicotinate-nucleotide-2 dimethylbenzimidazole phosphoribosyltransferase from3 methanocaldococcus jannaschii dsm 2661
|
|
|
|
| 7 | c3u4gA_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:namn:dmb phosphoribosyltransferase;
PDBTitle: the structure of cobt from pyrococcus horikoshii
|
|
|
|
| 8 | c4fwvA_
|
|
 |
87.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:ttc1975 peptidase;
PDBTitle: crystal structure of the n-terminal domain of the lon-like protease2 mtalonc
|
|
|
|
| 9 | d1htwa_
|
|
 |
63.4 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:YjeE-like |
|
|
|
| 10 | c6fpeE_
|
|
 |
48.2 |
17 |
PDB header:rna binding protein
Chain: E: PDB Molecule:atpase yjee, predicted to have essential role in cell wall
PDBTitle: bacterial protein complex
|
|
|
|
| 11 | c6qelL_
|
|
 |
46.3 |
20 |
PDB header:replication
Chain: L: PDB Molecule:dna replication protein dnac;
PDBTitle: e. coli dnabc apo complex
|
|
|
|
| 12 | c5c3cB_
|
|
 |
46.0 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:cbbq/nirq/norq domain protein;
PDBTitle: structural characterization of a newly identified component of alpha-2 carboxysomes: the aaa+ domain protein cso-cbbq
|
|
|
|
| 13 | c2zktB_
|
|
 |
43.4 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:2,3-bisphosphoglycerate-independent phosphoglycerate
PDBTitle: structure of ph0037 protein from pyrococcus horikoshii
|
|
|
|
| 14 | c5mvrA_
|
|
 |
41.6 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:trna threonylcarbamoyladenosine biosynthesis protein tsae;
PDBTitle: crystal structure of bacillus subtilus ydib
|
|
|
|
| 15 | c2c99A_
|
|
 |
38.5 |
11 |
PDB header:transcription regulation
Chain: A: PDB Molecule:psp operon transcriptional activator;
PDBTitle: structural basis of the nucleotide driven conformational2 changes in the aaa domain of transcription activator pspf
|
|
|
|
| 16 | c4ww4A_
|
|
 |
35.9 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:ruvb-like 1;
PDBTitle: double-heterohexameric rings of full-length rvb1(adp)/rvb2(adp)
|
|
|
|
| 17 | c4gvgB_
|
|
 |
35.5 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of salmonella typhimurium family 3 glycoside2 hydrolase (nagz)
|
|
|
|
| 18 | c2re2A_
|
|
 |
35.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein ta1041;
PDBTitle: crystal structure of a putative iron-molybdenum cofactor (femo-co)2 dinitrogenase (ta1041m) from thermoplasma acidophilum dsm 1728 at3 1.30 a resolution
|
|
|
|
| 19 | d1g41a_
|
|
 |
34.8 |
27 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
|
| 20 | c5bq5A_
|
|
 |
33.6 |
18 |
PDB header:atp-binding protein
Chain: A: PDB Molecule:insertion sequence is5376 putative atp-binding protein;
PDBTitle: crystal structure of the istb aaa+ domain bound to adp-bef3
|
|
|
|
| 21 | c3b5qB_ |
|
not modelled |
32.8 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of a putative sulfatase (np_810509.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.40 a resolution
|
|
|
| 22 | c3tqfA_ |
|
not modelled |
31.5 |
25 |
PDB header:transferase, hydrolase
Chain: A: PDB Molecule:hpr(ser) kinase;
PDBTitle: structure of the hpr(ser) kinase/phosphatase from coxiella burnetii
|
|
|
| 23 | c6qi8E_ |
|
not modelled |
31.4 |
31 |
PDB header:chaperone
Chain: E: PDB Molecule:ruvb-like 2;
PDBTitle: truncated human r2tp complex, structure 3 (adp-filled)
|
|
|
| 24 | c5he8J_ |
|
not modelled |
29.5 |
18 |
PDB header:protein binding
Chain: J: PDB Molecule:helicase loader;
PDBTitle: bacterial initiation protein
|
|
|
| 25 | c4uopB_ |
|
not modelled |
28.2 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:lipoteichoic acid primase;
PDBTitle: crystal structure of the lipoteichoic acid synthase ltap from listeria2 monocytogenes
|
|
|
| 26 | c5k4pA_ |
|
not modelled |
27.5 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:probable phosphatidylethanolamine transferase mcr-1;
PDBTitle: catalytic domain of mcr-1 phosphoethanolamine transferase
|
|
|
| 27 | c3ed4A_ |
|
not modelled |
27.1 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:arylsulfatase;
PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
|
|
|
| 28 | c2c9oC_ |
|
not modelled |
27.0 |
24 |
PDB header:hydrolase
Chain: C: PDB Molecule:ruvb-like 1;
PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
|
|
|
| 29 | d1g6oa_ |
|
not modelled |
26.9 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:RecA protein-like (ATPase-domain) |
|
|
| 30 | c3k6jA_ |
|
not modelled |
26.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein f01g10.3, confirmed by transcript evidence;
PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
|
|
|
| 31 | d1kkma_ |
|
not modelled |
25.9 |
42 |
Fold:PEP carboxykinase-like
Superfamily:PEP carboxykinase-like
Family:HPr kinase HprK C-terminal domain |
|
|
| 32 | c6c02B_ |
|
not modelled |
25.8 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 3 (enpp3,2 npp3, cd203c), inactive (t205a), n594s, with alpha,beta-methylene-atp3 (ampcpp)
|
|
|
| 33 | c4ug4H_ |
|
not modelled |
25.1 |
17 |
PDB header:hydrolase
Chain: H: PDB Molecule:choline sulfatase;
PDBTitle: crystal structure of a choline sulfatase from sinorhizobium2 melliloti
|
|
|
| 34 | d2tpta2 |
|
not modelled |
25.0 |
21 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 35 | c2qmhK_ |
|
not modelled |
24.9 |
42 |
PDB header:transferase
Chain: K: PDB Molecule:hpr kinase/phosphorylase;
PDBTitle: structure of v267f mutant hprk/p
|
|
|
| 36 | d1hdha_ |
|
not modelled |
24.8 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 37 | c5gz4A_ |
|
not modelled |
24.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:snake venom phosphodiesterase (pde);
PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra)
|
|
|
| 38 | c1ojlF_ |
|
not modelled |
24.7 |
27 |
PDB header:response regulator
Chain: F: PDB Molecule:transcriptional regulatory protein zrar;
PDBTitle: crystal structure of a sigma54-activator suggests the mechanism for2 the conformational switch necessary for sigma54 binding
|
|
|
| 39 | d1ko7a2 |
|
not modelled |
24.5 |
42 |
Fold:PEP carboxykinase-like
Superfamily:PEP carboxykinase-like
Family:HPr kinase HprK C-terminal domain |
|
|
| 40 | c4upiA_ |
|
not modelled |
23.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfatase family protein;
PDBTitle: dimeric sulfatase spas1 from silicibacter pomeroyi
|
|
|
| 41 | c2qzuA_ |
|
not modelled |
23.8 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative sulfatase yidj;
PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
|
|
|
| 42 | c4uorK_ |
|
not modelled |
23.6 |
19 |
PDB header:transferase
Chain: K: PDB Molecule:lipoteichoic acid synthase;
PDBTitle: structure of lipoteichoic acid synthase ltas from listeria2 monocytogenes in complex with glycerol phosphate
|
|
|
| 43 | c3rb8A_ |
|
not modelled |
23.5 |
9 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of the phage tubulin phuz(semet)-gdp
|
|
|
| 44 | c2w8dB_ |
|
not modelled |
23.3 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:processed glycerol phosphate lipoteichoic acid synthase 2;
PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
|
|
|
| 45 | c4uplC_ |
|
not modelled |
23.1 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:sulfatase family protein;
PDBTitle: dimeric sulfatase spas2 from silicibacter pomeroyi
|
|
|
| 46 | c5i5fA_ |
|
not modelled |
23.0 |
12 |
PDB header:membrane protein
Chain: A: PDB Molecule:inner membrane protein yejm;
PDBTitle: salmonella global domain 191
|
|
|
| 47 | c6b8vA_ |
|
not modelled |
23.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:adenylylsulfate kinase;
PDBTitle: crystal structure of adenylyl-sulfate kinase from cryptococcus2 neoformans
|
|
|
| 48 | c3lmaC_ |
|
not modelled |
22.8 |
23 |
PDB header:membrane protein
Chain: C: PDB Molecule:stage v sporulation protein ad (spovad);
PDBTitle: crystal structure of the stage v sporulation protein ad (spovad) from2 bacillus licheniformis. northeast structural genomics consortium3 target bir6.
|
|
|
| 49 | c5u9zB_ |
|
not modelled |
22.5 |
47 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoglycerol transferase;
PDBTitle: phosphoglycerol transferase gach from streptococcus pyogenes
|
|
|
| 50 | c5fqlA_ |
|
not modelled |
22.5 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:iduronate-2-sulfatase;
PDBTitle: insights into hunter syndrome from the structure of iduronate-2-2 sulfatase
|
|
|
| 51 | c6a82A_ |
|
not modelled |
22.2 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoethanolamine transferase eptc;
PDBTitle: crystal structure of the c-terminal periplasmic domain of eceptc from2 escherichia coli
|
|
|
| 52 | c4fdiA_ |
|
not modelled |
22.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylgalactosamine-6-sulfatase;
PDBTitle: the molecular basis of mucopolysaccharidosis iv a
|
|
|
| 53 | c4b56A_ |
|
not modelled |
21.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: structure of ectonucleotide pyrophosphatase-phosphodiesterase-12 (npp1)
|
|
|
| 54 | d2fywa1 |
|
not modelled |
21.7 |
38 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 55 | d1l8qa2 |
|
not modelled |
21.6 |
14 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
| 56 | c3co5B_ |
|
not modelled |
21.3 |
19 |
PDB header:transcription regulator
Chain: B: PDB Molecule:putative two-component system transcriptional response
PDBTitle: crystal structure of sigma-54 interaction domain of putative2 transcriptional response regulator from neisseria gonorrhoeae
|
|
|
| 57 | d1p49a_ |
|
not modelled |
21.3 |
14 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 58 | c3gh1A_ |
|
not modelled |
21.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted nucleotide-binding protein;
PDBTitle: crystal structure of predicted nucleotide-binding protein from vibrio2 cholerae
|
|
|
| 59 | c3bq9A_ |
|
not modelled |
21.0 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
|
|
| 60 | c3fdiA_ |
|
not modelled |
20.8 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of uncharacterized protein from eubacterium2 ventriosum atcc 27560.
|
|
|
| 61 | c5ep4A_ |
|
not modelled |
20.7 |
22 |
PDB header:transcription
Chain: A: PDB Molecule:putative repressor protein luxo;
PDBTitle: structure, regulation, and inhibition of the quorum-sensing signal2 integrator luxo
|
|
|
| 62 | c2gx8B_ |
|
not modelled |
20.7 |
33 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:nif3-related protein;
PDBTitle: the crystal structure of bacillus cereus protein related to nif3
|
|
|
| 63 | c2vhiG_ |
|
not modelled |
20.5 |
12 |
PDB header:hydrolase
Chain: G: PDB Molecule:cg3027-pa;
PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
|
|
|
| 64 | c5exsA_ |
|
not modelled |
20.4 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator fleq;
PDBTitle: aaa+ atpase fleq from pseudomonas aeruginosa bound to atp-gamma-s
|
|
|
| 65 | c2gzaB_ |
|
not modelled |
20.3 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:type iv secretion system protein virb11;
PDBTitle: crystal structure of the virb11 atpase from the brucella suis type iv2 secretion system in complex with sulphate
|
|
|
| 66 | c4ii7D_ |
|
not modelled |
20.3 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:flai atpase;
PDBTitle: archaellum assembly atpase flai
|
|
|
| 67 | d1fsua_ |
|
not modelled |
20.2 |
17 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 68 | c5tj3A_ |
|
not modelled |
20.1 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline phosphatase pafa;
PDBTitle: crystal structure of wild type alkaline phosphatase pafa to 1.7a2 resolution
|
|
|
| 69 | c5a22A_ |
|
not modelled |
19.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:vesicular stomatitis virus l polymerase;
PDBTitle: structure of the l protein of vesicular stomatitis virus from electron2 cryomicroscopy
|
|
|
| 70 | c2i09A_ |
|
not modelled |
19.7 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphopentomutase;
PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
|
|
|
| 71 | c4fxsA_ |
|
not modelled |
19.4 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 72 | c2we7A_ |
|
not modelled |
19.3 |
41 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of mycobacterium tuberculosis rv0376c2 homologue from mycobacterium smegmatis
|
|
|
| 73 | d1byia_ |
|
not modelled |
18.8 |
30 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
|
|
| 74 | c6b1vB_ |
|
not modelled |
18.8 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:iota-carrageenan sulfatase;
PDBTitle: crystal structure of ps i-cgsb c78s in complex with i-neocarratetraose
|
|
|
| 75 | c2c9oA_ |
|
not modelled |
18.6 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:ruvb-like 1;
PDBTitle: 3d structure of the human ruvb-like helicase ruvbl1
|
|
|
| 76 | c5gz5A_ |
|
not modelled |
18.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:snake venom phosphodiesterase (pde);
PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra) in complex with amp
|
|
|
| 77 | c6gfmA_ |
|
not modelled |
18.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrimidine/purine nucleotide 5'-monophosphate nucleosidase;
PDBTitle: crystal structure of the escherichia coli nucleosidase ppnn (pppgpp-2 form)
|
|
|
| 78 | d2gx8a1 |
|
not modelled |
17.8 |
33 |
Fold:NIF3 (NGG1p interacting factor 3)-like
Superfamily:NIF3 (NGG1p interacting factor 3)-like
Family:NIF3 (NGG1p interacting factor 3)-like |
|
|
| 79 | c5t4oA_ |
|
not modelled |
17.4 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase subunit alpha;
PDBTitle: autoinhibited e. coli atp synthase state 1
|
|
|
| 80 | c6blbA_ |
|
not modelled |
17.4 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:holliday junction atp-dependent dna helicase ruvb;
PDBTitle: 1.88 angstrom resolution crystal structure holliday junction atp-2 dependent dna helicase (ruvb) from pseudomonas aeruginosa in complex3 with adp
|
|
|
| 81 | c3h5qA_ |
|
not modelled |
17.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside phosphorylase;
PDBTitle: crystal structure of a putative pyrimidine-nucleoside phosphorylase2 from staphylococcus aureus
|
|
|
| 82 | c2dsjA_ |
|
not modelled |
17.0 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pyrimidine-nucleoside (thymidine) phosphorylase;
PDBTitle: crystal structure of project id tt0128 from thermus thermophilus hb8
|
|
|
| 83 | c4fw9A_ |
|
not modelled |
16.9 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:ttc1975 peptidase;
PDBTitle: crystal structure of the lon-like protease mtalonc
|
|
|
| 84 | d1knxa2 |
|
not modelled |
16.8 |
35 |
Fold:PEP carboxykinase-like
Superfamily:PEP carboxykinase-like
Family:HPr kinase HprK C-terminal domain |
|
|
| 85 | c1j6uA_ |
|
not modelled |
16.8 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
|
|
|
| 86 | c4uphA_ |
|
not modelled |
16.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfatase (sulfuric ester hydrolase) protein;
PDBTitle: crystal structure of phosphonate monoester hydrolase of agrobacterium2 radiobacter
|
|
|
| 87 | c4ga5H_ |
|
not modelled |
16.4 |
16 |
PDB header:transferase
Chain: H: PDB Molecule:putative thymidine phosphorylase;
PDBTitle: crystal structure of amp phosphorylase c-terminal deletion mutant in2 the apo-form
|
|
|
| 88 | c6hr5A_ |
|
not modelled |
16.1 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha-l-rhamnosidase/sulfatase (gh78);
PDBTitle: structure of the s1_25 family sulfatase module of the rhamnosidase2 fa22250 from formosa agariphila
|
|
|
| 89 | c2xrgA_ |
|
not modelled |
16.1 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
|
|
|
| 90 | d1qvra2 |
|
not modelled |
15.8 |
15 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
| 91 | c2xr9A_ |
|
not modelled |
15.7 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleotide pyrophosphatase/phosphodiesterase family
PDBTitle: crystal structure of autotaxin (enpp2)
|
|
|
| 92 | d1auka_ |
|
not modelled |
15.5 |
22 |
Fold:Alkaline phosphatase-like
Superfamily:Alkaline phosphatase-like
Family:Arylsulfatase |
|
|
| 93 | c1hqcB_ |
|
not modelled |
15.3 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:ruvb;
PDBTitle: structure of ruvb from thermus thermophilus hb8
|
|
|
| 94 | c5f4hF_ |
|
not modelled |
15.2 |
26 |
PDB header:hydrolase
Chain: F: PDB Molecule:nucleotide binding protein pinc;
PDBTitle: archael ruvb-like holiday junction helicase
|
|
|
| 95 | c3pfiB_ |
|
not modelled |
15.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:holliday junction atp-dependent dna helicase ruvb;
PDBTitle: 2.7 angstrom resolution crystal structure of a probable holliday2 junction dna helicase (ruvb) from campylobacter jejuni subsp. jejuni3 nctc 11168 in complex with adenosine-5'-diphosphate
|
|
|
| 96 | d1ixsb2 |
|
not modelled |
15.0 |
24 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
| 97 | c4xgcA_ |
|
not modelled |
14.9 |
20 |
PDB header:dna binding protein
Chain: A: PDB Molecule:origin recognition complex subunit 1;
PDBTitle: crystal structure of the eukaryotic origin recognition complex
|
|
|
| 98 | d1d2na_ |
|
not modelled |
14.9 |
18 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
|
|
| 99 | c1otpA_ |
|
not modelled |
14.8 |
19 |
PDB header:phosphorylase
Chain: A: PDB Molecule:thymidine phosphorylase;
PDBTitle: structural and theoretical studies suggest domain movement produces an2 active conformation of thymidine phosphorylase
|
|
|