| 1 |
|
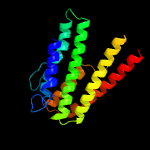
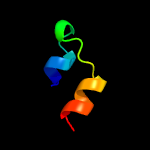
PDB 1m0f chain B
Region: 213 - 226
Aligned: 14
Modelled: 14
Confidence: 31.9%
Identity: 50%
PDB header:virus
Chain: B: PDB Molecule:scaffolding protein b;
PDBTitle: structural studies of bacteriophage alpha3 assembly, cryo-electron2 microscopy
Phyre2
| 2 |
|
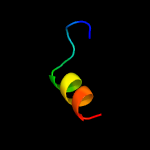
PDB 2v1s chain D
Region: 72 - 105
Aligned: 34
Modelled: 34
Confidence: 24.0%
Identity: 24%
PDB header:oxidoreductase
Chain: D: PDB Molecule:mitochondrial import receptor subunit tom20 homolog;
PDBTitle: crystal structure of rat tom20-aldh presequence complex
Phyre2
| 3 |
|
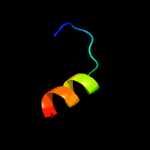
PDB 1om2 chain A
Region: 72 - 105
Aligned: 34
Modelled: 34
Confidence: 16.8%
Identity: 24%
Fold: Open three-helical up-and-down bundle
Superfamily: Mitochondrial import receptor subunit Tom20
Family: Mitochondrial import receptor subunit Tom20
Phyre2
| 4 |
|
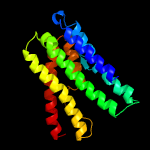
PDB 1pvg chain A domain 1
Region: 75 - 84
Aligned: 10
Modelled: 10
Confidence: 16.1%
Identity: 30%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 5 |
|
PDB 2d6f chain C domain 1
Region: 94 - 104
Aligned: 11
Modelled: 11
Confidence: 13.1%
Identity: 36%
Fold: GatB/YqeY motif
Superfamily: GatB/YqeY motif
Family: GatB/GatE C-terminal domain-like
Phyre2
| 6 |
|
PDB 5z7c chain A
Region: 81 - 110
Aligned: 30
Modelled: 30
Confidence: 11.1%
Identity: 30%
PDB header:metal binding protein
Chain: A: PDB Molecule:3'3'-cgamp-specific phosphodiesterase 3;
PDBTitle: crystal structure of cyclic gmp-amp specifc phosphodiesterases in2 v.cholerae (v-cgap3)
Phyre2
| 7 |
|
PDB 1otf chain A
Region: 89 - 104
Aligned: 16
Modelled: 16
Confidence: 10.3%
Identity: 6%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2
| 8 |
|
PDB 1kij chain A domain 1
Region: 75 - 84
Aligned: 10
Modelled: 10
Confidence: 9.9%
Identity: 20%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 9 |
|
PDB 1s16 chain A domain 1
Region: 75 - 84
Aligned: 10
Modelled: 10
Confidence: 9.6%
Identity: 30%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 10 |
|
PDB 1ei1 chain A domain 1
Region: 75 - 84
Aligned: 10
Modelled: 10
Confidence: 9.5%
Identity: 30%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: DNA gyrase/MutL, second domain
Phyre2
| 11 |
|
PDB 5k8z chain B
Region: 94 - 111
Aligned: 18
Modelled: 18
Confidence: 9.3%
Identity: 6%
PDB header:oxidoreductase
Chain: B: PDB Molecule:chlorite dismutase;
PDBTitle: crystal structure of dimeric chlorite dismutase from cyanothece sp.2 pcc7425 (ph 8.5)
Phyre2
| 12 |
|
PDB 6bd4 chain A
Region: 17 - 135
Aligned: 117
Modelled: 119
Confidence: 8.3%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:frizzled-4/rubredoxin chimeric protein;
PDBTitle: crystal structure of human apo-frizzled4 receptor
Phyre2
| 13 |
|
PDB 1ckq chain A
Region: 90 - 104
Aligned: 15
Modelled: 15
Confidence: 8.3%
Identity: 20%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease EcoRI
Phyre2
| 14 |
|
PDB 4r8z chain B
Region: 81 - 104
Aligned: 24
Modelled: 24
Confidence: 7.5%
Identity: 21%
PDB header:hydrolase
Chain: B: PDB Molecule:cyclic di-gmp phosphodiesterase;
PDBTitle: crystal structure of pa4781 hd-gyp domain from pseudomonas aeruginosa2 at 2.2a resolution showing a bi-metallic ni ion center
Phyre2
| 15 |
|
PDB 5d92 chain B
Region: 33 - 219
Aligned: 167
Modelled: 183
Confidence: 7.4%
Identity: 17%
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
Phyre2
| 16 |
|
PDB 4faz chain B
Region: 89 - 104
Aligned: 16
Modelled: 16
Confidence: 7.3%
Identity: 19%
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate isomerase protein;
PDBTitle: kinetic and structural characterization of the 4-oxalocrotonate2 tautomerase isozymes from methylibium petroleiphilum
Phyre2
| 17 |
|
PDB 1bjp chain A
Region: 89 - 104
Aligned: 16
Modelled: 16
Confidence: 6.9%
Identity: 6%
Fold: Tautomerase/MIF
Superfamily: Tautomerase/MIF
Family: 4-oxalocrotonate tautomerase-like
Phyre2
| 18 |
|
PDB 3ry0 chain A
Region: 89 - 104
Aligned: 16
Modelled: 16
Confidence: 6.5%
Identity: 25%
PDB header:isomerase
Chain: A: PDB Molecule:putative tautomerase;
PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
Phyre2
| 19 |
|
PDB 4o6m chain A
Region: 33 - 215
Aligned: 165
Modelled: 183
Confidence: 6.0%
Identity: 13%
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
Phyre2
| 20 |
|
PDB 4ef4 chain B
Region: 82 - 103
Aligned: 22
Modelled: 22
Confidence: 5.9%
Identity: 5%
PDB header:immune system
Chain: B: PDB Molecule:transmembrane protein 173;
PDBTitle: crystal structure of sting ctd complex with c-di-gmp
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|