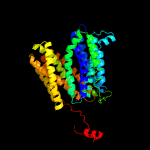
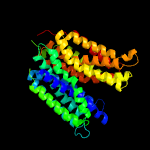
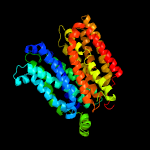
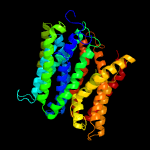
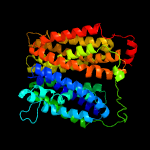
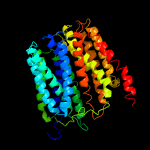
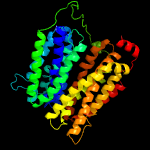
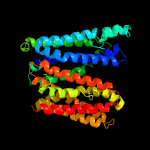
1 d1pw4a_
100.0
10
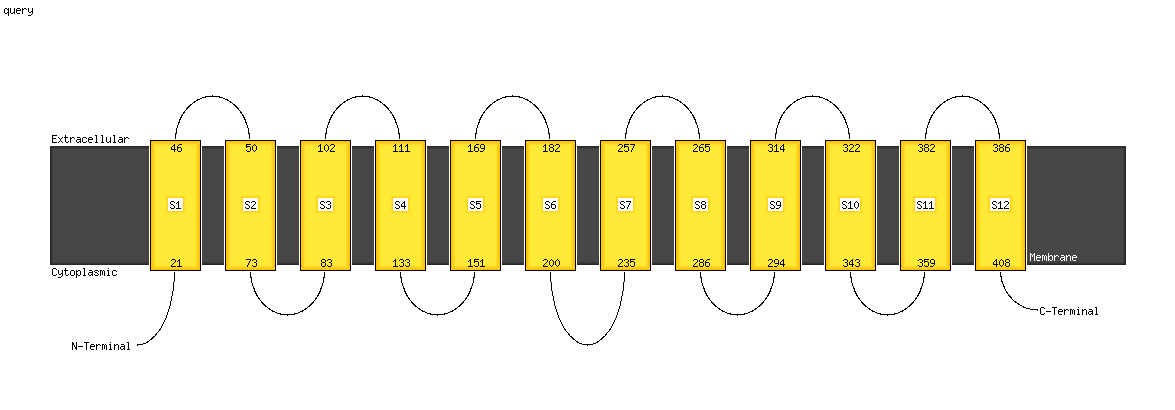
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: Glycerol-3-phosphate transporter
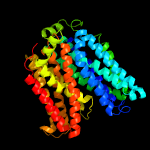
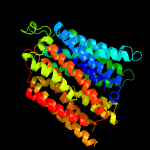
2 c4zp0A_
100.0
11
PDB header: transport proteinChain: A: PDB Molecule: multidrug transporter mdfa;PDBTitle: crystal structure of e. coli multidrug transporter mdfa in complex2 with deoxycholate
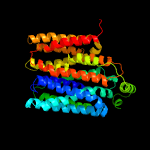
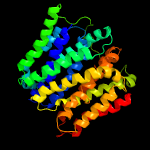
3 c3wdoA_
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: mfs transporter;PDBTitle: structure of e. coli yajr transporter
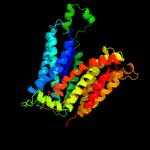
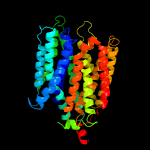
4 c6e9oA_
100.0
13
PDB header: membrane proteinChain: A: PDB Molecule: d-galactonate transport;PDBTitle: e. coli d-galactonate:proton symporter mutant e133q in the outward2 substrate-bound form
5 c4ldsB_
100.0
13
PDB header: transport protein, membrane proteinChain: B: PDB Molecule: bicyclomycin resistance protein tcab;PDBTitle: the inward-facing structure of the glucose transporter from2 staphylococcus epidermidis
6 c3o7pA_
100.0
10
PDB header: transport proteinChain: A: PDB Molecule: l-fucose-proton symporter;PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
7 c6g9xB_
100.0
9
PDB header: membrane proteinChain: B: PDB Molecule: major facilitator superfamily mfs_1;PDBTitle: crystal structure of a mfs transporter at 2.54 angstroem resolution
8 c6h7dA_
100.0
14
PDB header: membrane proteinChain: A: PDB Molecule: sugar transport protein 10;PDBTitle: crystal structure of a. thaliana sugar transport protein 10 in complex2 with glucose in the outward occluded state
9 c2gfpA_
100.0
14
PDB header: membrane proteinChain: A: PDB Molecule: multidrug resistance protein d;PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
10 c4j05A_
100.0
12
PDB header: transport proteinChain: A: PDB Molecule: phosphate transporter;PDBTitle: crystal structure of a eukaryotic phosphate transporter
11 c5aynA_
100.0
12
PDB header: transport proteinChain: A: PDB Molecule: solute carrier family 39 (iron-regulated transporter);PDBTitle: crystal structure of a bacterial homologue of iron transporter2 ferroportin in outward-facing state
12 c4apsB_
100.0
9
PDB header: transport proteinChain: B: PDB Molecule: di-or tripeptide h+ symporter;PDBTitle: crystal structure of a pot family peptide transporter in an inward2 open conformation.
13 c4ikyA_
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: di-tripeptide abc transporter (permease);PDBTitle: crystal structure of peptide transporter pot (e310q mutant) in complex2 with sulfate
14 c4ybqB_
100.0
14
PDB header: transport protein/immune systemChain: B: PDB Molecule: solute carrier family 2, facilitated glucose transporterPDBTitle: rat glut5 with fv in the outward-open form
15 c5c65A_
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: solute carrier family 2, facilitated glucose transporterPDBTitle: structure of the human glucose transporter glut3 / slc2a3
16 c6exsA_
100.0
11
PDB header: membrane proteinChain: A: PDB Molecule: peptide abc transporter permease;PDBTitle: crystal structure of a pot family transporter in complex with2 thioalcohol conjugated peptide.
17 d1pv7a_
100.0
12
Fold: MFS general substrate transporterSuperfamily: MFS general substrate transporterFamily: LacY-like proton/sugar symporter
18 c4cl5B_
100.0
8
PDB header: transport proteinChain: B: PDB Molecule: nitrate transporter 1.1;PDBTitle: crystal structure of the nitrate transporter nrt1.1 from2 arabidopsis thaliana in complex with nitrate.
19 c1pv7B_
100.0
12
PDB header: transport proteinChain: B: PDB Molecule: lactose permease;PDBTitle: crystal structure of lactose permease with tdg
20 c5aymA_
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: solute carrier family 39 (iron-regulated transporter);PDBTitle: crystal structure of a bacterial homologue of iron transporter2 ferroportin in outward-facing state with soaked iron
21 c6gs7A_
not modelled
100.0
12
PDB header: membrane proteinChain: A: PDB Molecule: dipeptide and tripeptide permease a;PDBTitle: crystal structure of peptide transporter dtpa-nanobody in glycine2 buffer
22 c4pypA_
not modelled
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: solute carrier family 2, facilitated glucose transporterPDBTitle: crystal structure of the human glucose transporter glut1
23 c6ei3A_
not modelled
100.0
13
PDB header: membrane proteinChain: A: PDB Molecule: proton-dependent oligopeptide transporter family protein;PDBTitle: crystal structure of auto inhibited pot family peptide transporter
24 c4q65A_
not modelled
100.0
12
PDB header: transport proteinChain: A: PDB Molecule: dipeptide permease d;PDBTitle: structure of the e. coli peptide transporter ybgh
25 c4w6vA_
not modelled
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: di-/tripeptide transporter;PDBTitle: crystal structure of a peptide transporter from yersinia2 enterocolitica at 3 a resolution
26 c4gbzA_
not modelled
100.0
15
PDB header: transport proteinChain: A: PDB Molecule: d-xylose-proton symporter;PDBTitle: the structure of the mfs (major facilitator superfamily) proton:xylose2 symporter xyle bound to d-glucose
27 c2xutC_
not modelled
100.0
12
PDB header: transport proteinChain: C: PDB Molecule: proton/peptide symporter family protein;PDBTitle: crystal structure of a proton dependent oligopeptide (pot) family2 transporter.
28 c4lepB_
not modelled
99.9
12
PDB header: membrane protein, tranport proteinChain: B: PDB Molecule: proton:oligopeptide symporter pot family;PDBTitle: structural insights into substrate recognition in proton dependent2 oligopeptide transporters
29 c4iu9A_
not modelled
99.9
12
PDB header: transport proteinChain: A: PDB Molecule: nitrite extrusion protein 2;PDBTitle: crystal structure of a membrane transporter
30 c4iu8A_
not modelled
99.9
12
PDB header: transport proteinChain: A: PDB Molecule: nitrite extrusion protein 2;PDBTitle: crystal structure of a membrane transporter (selenomethionine2 derivative)
31 c4m64D_
not modelled
99.8
7
PDB header: transport proteinChain: D: PDB Molecule: melibiose carrier protein;PDBTitle: 3d crystal structure of na+/melibiose symporter of salmonella2 typhimurium
32 c6ob7A_
not modelled
99.8
10
PDB header: transport proteinChain: A: PDB Molecule: equilibrative nucleoside transporter 1;PDBTitle: human equilibrative nucleoside transporter-1, dilazep bound
33 c5bz3A_
not modelled
30.5
14
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of sodium proton antiporter napa in outward-facing2 conformation.
34 c5gxwB_
not modelled
13.3
33
PDB header: protein bindingChain: B: PDB Molecule: peptide from nuclear mitotic apparatus protein 1;PDBTitle: importin and numa complex
35 c4bwzA_
not modelled
13.1
12
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of the sodium proton antiporter, napa
36 c2jmvA_
not modelled
12.2
21
PDB header: antiviral proteinChain: A: PDB Molecule: scytovirin;PDBTitle: solution structure of scytovirin refined against residual2 dipolar couplings
37 c2g9pA_
not modelled
10.1
14
PDB header: antimicrobial proteinChain: A: PDB Molecule: antimicrobial peptide latarcin 2a;PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
38 d2eiaa2
not modelled
9.4
36
Fold: Retrovirus capsid protein, N-terminal core domainSuperfamily: Retrovirus capsid protein, N-terminal core domainFamily: Retrovirus capsid protein, N-terminal core domain
39 c2kvtA_
not modelled
9.0
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yaia;PDBTitle: solution nmr structure of yaia from escherichia eoli. northeast2 structural genomics target er244
40 c3ukwC_
not modelled
8.4
33
PDB header: protein transport/inhibitorChain: C: PDB Molecule: bimax1 peptide;PDBTitle: mouse importin alpha: bimax1 peptide complex
41 c3lfmA_
not modelled
8.1
32
PDB header: oxidoreductaseChain: A: PDB Molecule: protein fto;PDBTitle: crystal structure of the fat mass and obesity associated (fto) protein2 reveals basis for its substrate specificity
42 c6cc4A_
not modelled
7.5
10
PDB header: transport proteinChain: A: PDB Molecule: soluble cytochrome b562, lipid ii flippase murj chimera;PDBTitle: structure of murj from escherichia coli
43 c5sxpG_
not modelled
7.3
33
PDB header: signaling protein/ligaseChain: G: PDB Molecule: e3 ubiquitin-protein ligase itchy homolog;PDBTitle: structural basis for the interaction between itch prr and beta-pix
44 c3vvpA_
not modelled
6.7
14
PDB header: transport proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of mate in complex with br-nrf
45 c5kzoA_
not modelled
6.6
24
PDB header: transcriptionChain: A: PDB Molecule: neurogenic locus notch homolog protein 1;PDBTitle: notch1 transmembrane and associated juxtamembrane segment
46 c5v2sA_
not modelled
6.2
17
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein b;PDBTitle: crystal structure of glycoprotein b from herpes simplex virus type i
47 c1rb8J_
not modelled
5.8
59
PDB header: virus/dnaChain: J: PDB Molecule: small core protein;PDBTitle: the phix174 dna binding protein j in two different capsid2 environments.