| 1 | c3dtfB_
|
|
 |
100.0 |
85 |
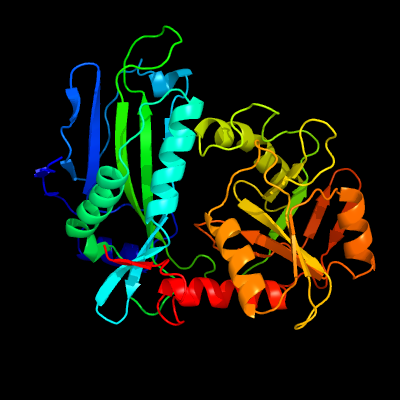
PDB header:transferase
Chain: B: PDB Molecule:branched-chain amino acid aminotransferase;
PDBTitle: structural analysis of mycobacterial branched chain aminotransferase-2 implications for inhibitor design
|
|
|
|
| 2 | d2a1ha1
|
|
 |
100.0 |
37 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
|
|
|
| 3 | c2abjG_
|
|
 |
100.0 |
37 |
PDB header:transferase
Chain: G: PDB Molecule:branched-chain-amino-acid aminotransferase, cytosolic;
PDBTitle: crystal structure of human branched chain amino acid transaminase in a2 complex with an inhibitor, c16h10n2o4f3scl, and pyridoxal 5'3 phosphate.
|
|
|
|
| 4 | c4dqnA_
|
|
 |
100.0 |
39 |
PDB header:transferase
Chain: A: PDB Molecule:putative branched-chain amino acid aminotransferase ilve;
PDBTitle: crystal structure of the branched-chain aminotransferase from2 streptococcus mutans
|
|
|
|
| 5 | c3uzbA_
|
|
 |
100.0 |
42 |
PDB header:transferase
Chain: A: PDB Molecule:branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structures of branched-chain aminotransferase from deinococcus2 radiodurans complexes with alpha-ketoisocaproate and l-glutamate3 suggest its radio-resistance for catalysis
|
|
|
|
| 6 | c3u0gA_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:putative branched-chain amino acid aminotransferase ilve;
PDBTitle: crystal structure of branched-chain amino acid aminotransferase from2 burkholderia pseudomallei
|
|
|
|
| 7 | c6h65A_
|
|
 |
100.0 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structure of the branched-chain-amino-acid aminotransferase2 from haliangium ochraceum
|
|
|
|
| 8 | d1iyea_
|
|
 |
100.0 |
34 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
|
|
|
| 9 | c6gkrC_
|
|
 |
100.0 |
29 |
PDB header:transferase
Chain: C: PDB Molecule:branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structure of branched-chain amino acid aminotransferase from2 thermobaculum terrenum in plp-form (holo-form)
|
|
|
|
| 10 | c6nstD_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: D: PDB Molecule:branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structure of branched chain amino acid aminotransferase from2 pseudomonas aeruginosa
|
|
|
|
| 11 | c1wrvB_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: B: PDB Molecule:branched-chain amino acid aminotransferase;
PDBTitle: crystal structure of t.th.hb8 branched-chain amino acid2 aminotransferase
|
|
|
|
| 12 | c5ce8B_
|
|
 |
100.0 |
30 |
PDB header:transferase
Chain: B: PDB Molecule:branched-chain amino acid aminotransferase;
PDBTitle: crystal structure of branched-chain aminotransferase from thermophilic2 archaea thermoproteus uzoniensis
|
|
|
|
| 13 | c3wwjE_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: E: PDB Molecule:(r)-amine transaminase;
PDBTitle: crystal structure of an engineered sitagliptin-producing transaminase,2 ata-117-rd11
|
|
|
|
| 14 | c4ce5A_
|
|
 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:at-omegata;
PDBTitle: first crystal structure of an (r)-selective omega-transaminase2 from aspergillus terreus
|
|
|
|
| 15 | c4m0jA_
|
|
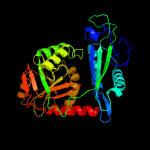 |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:d-amino acid aminotransferase;
PDBTitle: crystal structure of a d-amino acid aminotransferase from burkholderia2 thailandensis e264
|
|
|
|
| 16 | c3cswB_
|
|
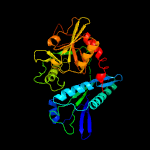 |
100.0 |
27 |
PDB header:transferase
Chain: B: PDB Molecule:putative branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structure of a putative branched-chain amino acid2 aminotransferase (tm0831) from thermotoga maritima at 2.15 a3 resolution
|
|
|
|
| 17 | c5cm0A_
|
|
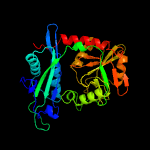 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:branched-chain transaminase;
PDBTitle: crystal structure of branched-chain aminotransferase from thermophilic2 archaea geoglobus acetivorans
|
|
|
|
| 18 | c4tviB_
|
|
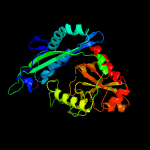 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:aminotransferase, class iv;
PDBTitle: x-ray crystal structure of an aminotransferase from brucella abortus2 bound to the co-factor plp
|
|
|
|
| 19 | d1daaa_
|
|
 |
100.0 |
22 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
|
|
|
| 20 | c2xpfB_
|
|
 |
100.0 |
29 |
PDB header:lyase
Chain: B: PDB Molecule:4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative aminodeoxychorismate lyase2 from pseudomonas aeruginosa
|
|
|
|
| 21 | c4jxuB_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:putative aminotransferase;
PDBTitle: structure of aminotransferase ilve2 from sinorhizobium meliloti2 complexed with plp
|
|
|
| 22 | c5k3wA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:cputa1;
PDBTitle: structural characterisation of fold iv-transaminase, cputa1, from2 curtobacterium pusillum
|
|
|
| 23 | d1i2ka_ |
|
not modelled |
100.0 |
19 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
|
|
| 24 | c6bb9A_ |
|
not modelled |
100.0 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:4-amino-4-deoxychorismate lyase;
PDBTitle: the crystal structure of 4-amino-4-deoxychorismate lyase from2 salmonella typhimurium lt2
|
|
|
| 25 | c3lulA_ |
|
not modelled |
100.0 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative 4-amino-4-deoxychorismate lyase.2 (yp_094631.1) from legionella pneumophila subsp. pneumophila str.3 philadelphia 1 at 1.78 a resolution
|
|
|
| 26 | c2zgiA_ |
|
not modelled |
100.0 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:putative 4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative 4-amino-4-deoxychorismate lyase
|
|
|
| 27 | c3snoA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical aminotransferase;
PDBTitle: crystal structure of a putative aminotransferase (ncgl2491) from2 corynebacterium glutamicum atcc 13032 at 1.60 a resolution
|
|
|
| 28 | c3qqmD_ |
|
not modelled |
100.0 |
26 |
PDB header:transferase
Chain: D: PDB Molecule:mlr3007 protein;
PDBTitle: crystal structure of a putative amino-acid aminotransferase2 (np_104211.1) from mesorhizobium loti at 2.30 a resolution
|
|
|
| 29 | c4k6nA_ |
|
not modelled |
100.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:aminodeoxychorismate lyase;
PDBTitle: crystal structure of yeast 4-amino-4-deoxychorismate lyase
|
|
|
| 30 | c3cebA_ |
|
not modelled |
100.0 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:d-aminoacid aminotransferase-like plp-dependent enzyme;
PDBTitle: crystal structure of a putative 4-amino-4-deoxychorismate lyase2 (hs_0128) from haemophilus somnus 129pt at 2.40 a resolution
|
|
|
| 31 | c4h5fB_ |
|
not modelled |
66.6 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:amino acid abc superfamily atp binding cassette
PDBTitle: crystal structure of an amino acid abc transporter substrate-binding2 protein from streptococcus pneumoniae canada mdr_19a bound to l-3 arginine, form 1
|
|
|
| 32 | c4q0cA_ |
|
not modelled |
65.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:virulence sensor protein bvgs;
PDBTitle: 3.1 a resolution crystal structure of the b. pertussis bvgs2 periplasmic domain
|
|
|
| 33 | c2ylnA_ |
|
not modelled |
60.6 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:putative abc transporter, periplasmic binding protein,
PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
|
|
|
| 34 | c5t0wA_ |
|
not modelled |
60.3 |
22 |
PDB header:transport protein
Chain: A: PDB Molecule:anccdt-1;
PDBTitle: crystal structure of the ancestral amino acid-binding protein anccdt-2 1, a precursor of cyclohexadienyl dehydratase
|
|
|
| 35 | c5gzsA_ |
|
not modelled |
60.3 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:ggdef family protein;
PDBTitle: structure of vc protein
|
|
|
| 36 | d1wdna_ |
|
not modelled |
58.0 |
17 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 37 | c4pshA_ |
|
not modelled |
55.9 |
14 |
PDB header:protein transport
Chain: A: PDB Molecule:abc-type transporter, periplasmic subunit family 3;
PDBTitle: structure of holo argbp from t. maritima
|
|
|
| 38 | c3k4uA_ |
|
not modelled |
55.8 |
12 |
PDB header:transport protein
Chain: A: PDB Molecule:binding component of abc transporter;
PDBTitle: crystal structure of putative binding component of abc transporter2 from wolinella succinogenes dsm 1740 complexed with lysine
|
|
|
| 39 | c4ymxB_ |
|
not modelled |
54.8 |
22 |
PDB header:transport protein
Chain: B: PDB Molecule:abc-type amino acid transport system, periplasmic
PDBTitle: crystal structure of the substrate binding protein of an amino acid2 abc transporter
|
|
|
| 40 | c3r39A_ |
|
not modelled |
54.5 |
17 |
PDB header:transport protein
Chain: A: PDB Molecule:putative periplasmic binding protein;
PDBTitle: crystal structure of periplasmic d-alanine abc transporter from2 salmonella enterica
|
|
|
| 41 | c3kzgB_ |
|
not modelled |
54.1 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:arginine 3rd transport system periplasmic binding
PDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
|
|
|
| 42 | c4f3pB_ |
|
not modelled |
53.9 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamine-binding periplasmic protein;
PDBTitle: crystal structure of a glutamine-binding periplasmic protein from2 burkholderia pseudomallei in complex with glutamine
|
|
|
| 43 | c3g41A_ |
|
not modelled |
51.4 |
16 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid abc transporter, periplasmic amino acid-binding
PDBTitle: the structure of cpn0482, the arginine binding protein from the2 periplasm of chlamydia pneumoniae
|
|
|
| 44 | c3vvfA_ |
|
not modelled |
49.1 |
18 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid abc transporter, binding protein;
PDBTitle: crystal structure of ttc0807 complexed with arginine
|
|
|
| 45 | c4pp0B_ |
|
not modelled |
48.4 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:nopaline-binding periplasmic protein;
PDBTitle: structure of the pbp noct-m117n in complex with pyronopaline
|
|
|
| 46 | d1hsla_ |
|
not modelled |
45.8 |
14 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 47 | c6dnmA_ |
|
not modelled |
45.7 |
11 |
PDB header:chaperone
Chain: A: PDB Molecule:export chaperone sats;
PDBTitle: the crystal structure of sats c-terminal domain
|
|
|
| 48 | c4hubI_ |
|
not modelled |
43.9 |
24 |
PDB header:ribosome
Chain: I: PDB Molecule:50s ribosomal protein l11p;
PDBTitle: the re-refined crystal structure of the haloarcula marismortui large2 ribosomal subunit at 2.4 angstrom resolution: more complete structure3 of the l7/l12 and l1 stalk, l5 and lx proteins
|
|
|
| 49 | c4eq9A_ |
|
not modelled |
43.8 |
9 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter substrate-binding protein-amino acid
PDBTitle: 1.4 angstrom crystal structure of abc transporter glutathione-binding2 protein gsht from streptococcus pneumoniae strain canada mdr_19a in3 complex with glutathione
|
|
|
| 50 | d1lsta_ |
|
not modelled |
43.6 |
12 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 51 | c4i62A_ |
|
not modelled |
42.2 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid abc transporter, periplasmic amino acid-binding
PDBTitle: 1.05 angstrom crystal structure of an amino acid abc transporter2 substrate-binding protein abpa from streptococcus pneumoniae canada3 mdr_19a bound to l-arginine
|
|
|
| 52 | c4oenA_ |
|
not modelled |
41.1 |
24 |
PDB header:transport protein
Chain: A: PDB Molecule:second substrate binding domain of putative amino acid abc
PDBTitle: crystal structure of the second substrate binding domain of a putative2 amino acid abc transporter from streptococcus pneumoniae canada3 mdr_19a
|
|
|
| 53 | c2ieeB_ |
|
not modelled |
40.9 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable abc transporter extracellular-binding protein
PDBTitle: crystal structure of yckb_bacsu from bacillus subtilis. northeast2 structural genomics consortium target sr574.
|
|
|
| 54 | c2y7iB_ |
|
not modelled |
39.1 |
10 |
PDB header:arginine-binding protein
Chain: B: PDB Molecule:stm4351;
PDBTitle: structural basis for high arginine specificity in salmonella2 typhimurium periplasmic binding protein stm4351.
|
|
|
| 55 | c5colB_ |
|
not modelled |
38.1 |
32 |
PDB header:translation
Chain: B: PDB Molecule:50s ribosomal protein l11;
PDBTitle: ribosomal protein l11 from methanococcus jannaschii
|
|
|
| 56 | c3h7mA_ |
|
not modelled |
36.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of a histidine kinase sensor domain with similarity2 to periplasmic binding proteins
|
|
|
| 57 | c4kptA_ |
|
not modelled |
35.7 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:glutamine abc transporter permease and substrate binding
PDBTitle: crystal structure of substrate binding domain 1 (sbd1) of abc2 transporter glnpq from lactococcus lactis
|
|
|
| 58 | c5orgA_ |
|
not modelled |
35.5 |
14 |
PDB header:octopine-binding protein
Chain: A: PDB Molecule:octopine-binding periplasmic protein;
PDBTitle: structure of the periplasmic binding protein (pbp) occj from a.2 tumefaciens b6 in complex with octopine.
|
|
|
| 59 | c4kr5B_ |
|
not modelled |
34.9 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamine abc transporter permease and substrate binding
PDBTitle: crystal structure of lactococcus lactis glnp substrate binding domain2 2 (sbd2) in open conformation
|
|
|
| 60 | c6detA_ |
|
not modelled |
34.7 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:tv2483;
PDBTitle: the crystal structure of tv2483 bound to l-arginine
|
|
|
| 61 | c6h2tA_ |
|
not modelled |
34.5 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:probable glutamine-binding lipoprotein glnh (glnbp);
PDBTitle: glnh bound to glu, mycobacterium tuberculosis
|
|
|
| 62 | c2q2aD_ |
|
not modelled |
32.3 |
20 |
PDB header:transport protein
Chain: D: PDB Molecule:artj;
PDBTitle: crystal structures of the arginine-, lysine-, histidine-binding2 protein artj from the thermophilic bacterium geobacillus3 stearothermophilus
|
|
|
| 63 | c4oz9A_ |
|
not modelled |
30.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:membrane-bound lytic murein transglycosylase f;
PDBTitle: crystal structure of mltf from pseudomonas aeruginosa complexed with2 isoleucine
|
|
|
| 64 | c5hmtA_ |
|
not modelled |
30.5 |
15 |
PDB header:transport protein
Chain: A: PDB Molecule:cyclohexadienyl dehydratase;
PDBTitle: crystal structure of the cyclohexadienyl dehydratase-like solute-2 binding protein sar11_1068 from candidatus pelagibacter ubique.
|
|
|
| 65 | c5josA_ |
|
not modelled |
29.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:cyclohexadienyl dehydratase;
PDBTitle: crystal structure of an ancestral cyclohexadienyl dehydratase, anccdt-2 3(p188l).
|
|
|
| 66 | c4g4pA_ |
|
not modelled |
28.9 |
23 |
PDB header:transport protein
Chain: A: PDB Molecule:amino acid abc transporter, amino acid-binding/permease
PDBTitle: crystal structure of glutamine-binding protein from enterococcus2 faecalis at 1.5 a
|
|
|
| 67 | d2f34a1 |
|
not modelled |
28.9 |
19 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 68 | c3hv1A_ |
|
not modelled |
28.6 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:polar amino acid abc uptake transporter substrate
PDBTitle: crystal structure of a polar amino acid abc uptake2 transporter substrate binding protein from streptococcus3 thermophilus
|
|
|
| 69 | c6a80B_ |
|
not modelled |
28.4 |
12 |
PDB header:transport protein
Chain: B: PDB Molecule:putative amino acid-binding periplasmic abc transporter
PDBTitle: crystal structure of putative amino acid binding periplasmic abc2 transporter protein from candidatus liberibacter asiaticus in complex3 with cystine
|
|
|
| 70 | c5dtbB_ |
|
not modelled |
28.2 |
15 |
PDB header:membrane protein
Chain: B: PDB Molecule:cg3822;
PDBTitle: crystal structure of the drosophila cg3822 kair1d ligand binding2 domain complex with glutamate
|
|
|
| 71 | c2hi1A_ |
|
not modelled |
27.2 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:4-hydroxythreonine-4-phosphate dehydrogenase 2;
PDBTitle: the structure of a putative 4-hydroxythreonine-4-phosphate2 dehydrogenase from salmonella typhimurium.
|
|
|
| 72 | c4ohnA_ |
|
not modelled |
27.1 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter substrate-binding protein;
PDBTitle: crystal structure of an abc uptake transporter substrate binding2 protein from streptococcus pneumoniae with bound histidine
|
|
|
| 73 | c5ikbA_ |
|
not modelled |
26.9 |
19 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate receptor ionotropic, kainate 4,glutamate receptor
PDBTitle: crystal structure of the kainate receptor gluk4 ligand binding domain2 in complex with kainate
|
|
|
| 74 | c3delC_ |
|
not modelled |
26.7 |
14 |
PDB header:protein binding, transport protein
Chain: C: PDB Molecule:arginine binding protein;
PDBTitle: the structure of ct381, the arginine binding protein from the2 periplasm chlamydia trachomatis
|
|
|
| 75 | c3chgB_ |
|
not modelled |
25.9 |
14 |
PDB header:ligand binding protein
Chain: B: PDB Molecule:glycine betaine-binding protein;
PDBTitle: the compatible solute-binding protein opuac from bacillus2 subtilis in complex with dmsa
|
|
|
| 76 | c2kngA_ |
|
not modelled |
25.7 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: solution structure of c-domain of lsr2
|
|
|
| 77 | c2pyyB_ |
|
not modelled |
24.4 |
17 |
PDB header:transport protein
Chain: B: PDB Molecule:ionotropic glutamate receptor bacterial homologue;
PDBTitle: crystal structure of the glur0 ligand-binding core from nostoc2 punctiforme in complex with (l)-glutamate
|
|
|
| 78 | d1vqon1 |
|
not modelled |
24.3 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
|
|
| 79 | c2o1mB_ |
|
not modelled |
23.4 |
8 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:probable amino-acid abc transporter extracellular-binding
PDBTitle: crystal structure of the probable amino-acid abc transporter2 extracellular-binding protein ytmk from bacillus subtilis. northeast3 structural genomics consortium target sr572
|
|
|
| 80 | d2ic1a1 |
|
not modelled |
23.3 |
12 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Cysteine dioxygenase type I |
|
|
| 81 | c6f37B_ |
|
not modelled |
22.9 |
14 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:nano3,fucose-binding lectin protein;
PDBTitle: fusion protein of rsl and trimeric coiled coil
|
|
|
| 82 | c4zv2A_ |
|
not modelled |
22.5 |
27 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:ancqr;
PDBTitle: an ancestral arginine-binding protein bound to glutamine
|
|
|
| 83 | c1vcnA_ |
|
not modelled |
22.1 |
34 |
PDB header:ligase
Chain: A: PDB Molecule:ctp synthetase;
PDBTitle: crystal structure of t.th. hb8 ctp synthetase complex with sulfate2 anion
|
|
|
| 84 | c3i6vA_ |
|
not modelled |
22.1 |
20 |
PDB header:transport protein
Chain: A: PDB Molecule:periplasmic his/glu/gln/arg/opine family-binding protein;
PDBTitle: crystal structure of a periplasmic his/glu/gln/arg/opine family-2 binding protein from silicibacter pomeroyi in complex with lysine
|
|
|
| 85 | c2xx7B_ |
|
not modelled |
21.6 |
20 |
PDB header:transport protein
Chain: B: PDB Molecule:glutamate receptor 2;
PDBTitle: crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-2 (trifluoromethyl)-4,5,6,7-tetrahydro-1h-indazole in complex with3 the ligand binding domain of the rat glua2 receptor and glutamate4 at 2.2a resolution.
|
|
|
| 86 | c2rejA_ |
|
not modelled |
21.5 |
8 |
PDB header:choline-binding protein
Chain: A: PDB Molecule:putative glycine betaine abc transporter protein;
PDBTitle: abc-transporter choline binding protein in unliganded semi-2 closed conformation
|
|
|
| 87 | c4z9nB_ |
|
not modelled |
21.2 |
16 |
PDB header:transport protein
Chain: B: PDB Molecule:amino acid abc transporter, periplasmic amino acid-binding
PDBTitle: abc transporter / periplasmic binding protein from brucella ovis with2 glutathione bound
|
|
|
| 88 | c3kbrA_ |
|
not modelled |
20.7 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:cyclohexadienyl dehydratase;
PDBTitle: the crystal structure of cyclohexadienyl dehydratase precursor from2 pseudomonas aeruginosa pa01
|
|
|
| 89 | c4qicB_ |
|
not modelled |
20.7 |
23 |
PDB header:signaling protein/dna binding protein
Chain: B: PDB Molecule:anti-sigma factor nepr;
PDBTitle: co-crystal structure of anti-anti-sigma factor phyr complexed with2 anti-sigma factor nepr from bartonella quintana
|
|
|
| 90 | d1mqia_ |
|
not modelled |
20.4 |
22 |
Fold:Periplasmic binding protein-like II
Superfamily:Periplasmic binding protein-like II
Family:Phosphate binding protein-like |
|
|
| 91 | d1dzfa1 |
|
not modelled |
20.0 |
7 |
Fold:Restriction endonuclease-like
Superfamily:Eukaryotic RPB5 N-terminal domain
Family:Eukaryotic RPB5 N-terminal domain |
|
|
| 92 | c5uh0A_ |
|
not modelled |
19.6 |
12 |
PDB header:hydrolase,oxidoreductase
Chain: A: PDB Molecule:membrane-bound lytic murein transglycosylase f;
PDBTitle: 1.95 angstrom resolution crystal structure of fragment (35-274) of2 membrane-bound lytic murein transglycosylase f from yersinia pestis.
|
|
|
| 93 | d1mxsa_ |
|
not modelled |
19.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 94 | d3elna1 |
|
not modelled |
18.9 |
13 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Cysteine dioxygenase type I |
|
|
| 95 | c4alzA_ |
|
not modelled |
18.9 |
16 |
PDB header:membrane protein
Chain: A: PDB Molecule:yop proteins translocation protein d;
PDBTitle: the yersinia t3ss basal body component yscd reveals a different2 structural periplasmatic domain organization to known homologue prgh
|
|
|
| 96 | c4c0rB_ |
|
not modelled |
18.3 |
6 |
PDB header:transport protein
Chain: B: PDB Molecule:putative amino acid binding protein;
PDBTitle: molecular and structural basis of glutathione import in2 gram-positive bacteria via gsht and the cystine abc3 importer tcybc of streptococcus mutans.
|
|
|
| 97 | c4zdmA_ |
|
not modelled |
18.3 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:glutamate receptor kainate-like protein;
PDBTitle: pleurobrachia bachei iglur3 lbd glycine complex
|
|
|
| 98 | c6gpcB_ |
|
not modelled |
18.0 |
14 |
PDB header:transport protein
Chain: B: PDB Molecule:amino acid abc transporter, periplasmic amino acid-binding
PDBTitle: crystal structure of the arginine-bound form of domain 1 from tmargbp
|
|
|
| 99 | d1uxca_ |
|
not modelled |
17.6 |
7 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:GalR/LacI-like bacterial regulator |
|
|