| 1 | c3k7dA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate-ammonia-ligase adenylyltransferase;
PDBTitle: c-terminal (adenylylation) domain of e.coli glutamine synthetase2 adenylyltransferase
|
|
|
|
| 2 | c1v4aA_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:glutamate-ammonia-ligase adenylyltransferase;
PDBTitle: structure of the n-terminal domain of escherichia coli2 glutamine synthetase adenylyltransferase
|
|
|
|
| 3 | d1v4aa2
|
|
 |
100.0 |
23 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:GlnE-like domain |
|
|
|
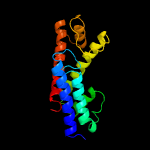
| 4 | d1v4aa1
|
|
 |
100.0 |
27 |
Fold:Four-helical up-and-down bundle
Superfamily:Nucleotidyltransferase substrate binding subunit/domain
Family:Glutamine synthase adenylyltransferase GlnE, domain 2 |
|
|
|
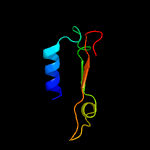
| 5 | d1knya2
|
|
 |
97.0 |
20 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Kanamycin nucleotidyltransferase (KNTase), N-terminal domain |
|
|
|
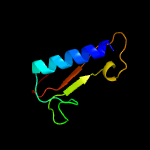
| 6 | c1knyA_
|
|
 |
96.5 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:kanamycin nucleotidyltransferase;
PDBTitle: kanamycin nucleotidyltransferase
|
|
|
|
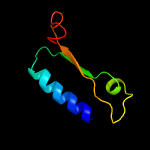
| 7 | c2rffA_
|
|
 |
96.3 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative nucleotidyltransferase;
PDBTitle: crystal structure of a putative nucleotidyltransferase (np_343093.1)2 from sulfolobus solfataricus at 1.40 a resolution
|
|
|
|
| 8 | d1ylqa1
|
|
 |
95.3 |
16 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
|
|
|
| 9 | d1no5a_
|
|
 |
94.3 |
20 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
|
|
|
| 10 | c4cs6A_
|
|
 |
93.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:aminoglycoside adenyltransferase;
PDBTitle: crystal structure of aada - an aminoglycoside adenyltransferase
|
|
|
|
| 11 | d1wota_
|
|
 |
93.5 |
24 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Catalytic subunit of bi-partite nucleotidyltransferase |
|
|
|
| 12 | c3c66B_
|
|
 |
92.8 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) polymerase;
PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
|
|
|
|
| 13 | d2q66a2
|
|
 |
92.1 |
15 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly(A) polymerase, PAP, N-terminal domain |
|
|
|
| 14 | c1q78A_
|
|
 |
92.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) polymerase alpha;
PDBTitle: crystal structure of poly(a) polymerase in complex with 3'-2 datp and magnesium chloride
|
|
|
|
| 15 | d1q79a2
|
|
 |
90.8 |
16 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Poly(A) polymerase, PAP, N-terminal domain |
|
|
|
| 16 | c4ebjB_
|
|
 |
90.5 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:aminoglycoside nucleotidyltransferase;
PDBTitle: crystal structure of aminoglycoside 4'-o-adenylyltransferase ant(4')-2 iib, apo
|
|
|
|
| 17 | c3jz0B_
|
|
 |
89.3 |
16 |
PDB header:transferase/antibiotic
Chain: B: PDB Molecule:lincosamide nucleotidyltransferase;
PDBTitle: linb complexed with clindamycin and ampcpp
|
|
|
|
| 18 | c5z4cA_
|
|
 |
85.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:terminal uridylyltransferase tailor;
PDBTitle: crystal structure of tailor
|
|
|
|
| 19 | d2fmpa3
|
|
 |
78.5 |
17 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
|
|
|
| 20 | c4zrlA_
|
|
 |
75.0 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) rna polymerase gld-2;
PDBTitle: structure of the non canonical poly(a) polymerase complex gld-2 - gld-2 3
|
|
|
|
| 21 | c2bcuA_ |
|
not modelled |
72.3 |
19 |
PDB header:transferase, lyase/dna
Chain: A: PDB Molecule:dna polymerase lambda;
PDBTitle: dna polymerase lambda in complex with a dna duplex2 containing an unpaired damp and a t:t mismatch
|
|
|
| 22 | c4xq7A_ |
|
not modelled |
68.2 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:2'-5'-oligoadenylate synthase-like protein;
PDBTitle: the crystal structure of the oas-like domain (old) of human oasl
|
|
|
| 23 | c5w0mA_ |
|
not modelled |
67.0 |
11 |
PDB header:transferase/rna
Chain: A: PDB Molecule:terminal uridylyltransferase 7;
PDBTitle: structure of human tut7 catalytic module (cm) in complex with u5 rna
|
|
|
| 24 | c4ep7A_ |
|
not modelled |
62.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) rna polymerase protein cid1;
PDBTitle: functional implications from the cid1 poly(u) polymerase crystal2 structure
|
|
|
| 25 | c4e80B_ |
|
not modelled |
61.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) rna polymerase protein cid1;
PDBTitle: structural basis for the activity of a cytoplasmic rna terminal u-2 transferase
|
|
|
| 26 | c4s3nA_ |
|
not modelled |
54.8 |
16 |
PDB header:transferase/rna
Chain: A: PDB Molecule:2'-5'-oligoadenylate synthase 3;
PDBTitle: crystal structure of human oas3 domain i in complex with dsrna
|
|
|
| 27 | d2vana2 |
|
not modelled |
54.3 |
17 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
|
|
| 28 | c2la3A_ |
|
not modelled |
53.9 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
|
|
|
| 29 | c4p4oA_ |
|
not modelled |
51.6 |
11 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase beta;
PDBTitle: crystal structure of leishmania infantum polymerase beta: ternary gap2 complex
|
|
|
| 30 | c4uz0B_ |
|
not modelled |
49.8 |
30 |
PDB header:apoptosis
Chain: B: PDB Molecule:nucleolar protein 3;
PDBTitle: crystal structure of apoptosis repressor with card (arc)
|
|
|
| 31 | c1sz1A_ |
|
not modelled |
49.0 |
16 |
PDB header:transferase/rna
Chain: A: PDB Molecule:trna nucleotidyltransferase;
PDBTitle: mechanism of cca-adding enzymes specificity revealed by crystal2 structures of ternary complexes
|
|
|
| 32 | c3nybA_ |
|
not modelled |
48.8 |
14 |
PDB header:transferase/rna binding protein
Chain: A: PDB Molecule:poly(a) rna polymerase protein 2;
PDBTitle: structure and function of the polymerase core of tramp, a rna2 surveillance complex
|
|
|
| 33 | c5yepA_ |
|
not modelled |
46.8 |
17 |
PDB header:antitoxin/toxin
Chain: A: PDB Molecule:toxin-antitoxin system antidote mnt family;
PDBTitle: crystal structure of so_3166-so_3165 from shewanella oneidensis
|
|
|
| 34 | d1jmsa4 |
|
not modelled |
39.7 |
13 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
|
|
| 35 | c5m45K_ |
|
not modelled |
38.6 |
25 |
PDB header:ligase
Chain: K: PDB Molecule:acetone carboxylase beta subunit;
PDBTitle: structure of acetone carboxylase purified from xanthobacter2 autotrophicus
|
|
|
| 36 | c1kdhA_ |
|
not modelled |
35.5 |
18 |
PDB header:transferase/dna
Chain: A: PDB Molecule:terminal deoxynucleotidyltransferase short
PDBTitle: binary complex of murine terminal deoxynucleotidyl2 transferase with a primer single stranded dna
|
|
|
| 37 | c3fioB_ |
|
not modelled |
35.4 |
7 |
PDB header:nucleotide binding protein, metal bindin
Chain: B: PDB Molecule:a cystathionine beta-synthase domain protein
PDBTitle: crystal structure of a fragment of a cystathionine beta-2 synthase domain protein fused to a zn-ribbon-like domain
|
|
|
| 38 | c1x4pA_ |
|
not modelled |
35.4 |
21 |
PDB header:rna binding protein
Chain: A: PDB Molecule:putative splicing factor, arginine/serine-rich
PDBTitle: solution structure of surp domain in sfrs14 protei
|
|
|
| 39 | d1r89a2 |
|
not modelled |
33.6 |
14 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:Archaeal tRNA CCA-adding enzyme catalytic domain |
|
|
| 40 | d1vk2a_ |
|
not modelled |
32.2 |
14 |
Fold:Uracil-DNA glycosylase-like
Superfamily:Uracil-DNA glycosylase-like
Family:Mug-like |
|
|
| 41 | c3db2C_ |
|
not modelled |
32.2 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
|
|
| 42 | c1tltB_ |
|
not modelled |
31.7 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase (virulence factor mvim homolog);
PDBTitle: crystal structure of a putative oxidoreductase (virulence factor mvim2 homolog)
|
|
|
| 43 | c5l9wb_ |
|
not modelled |
29.4 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 44 | d2bcqa3 |
|
not modelled |
28.3 |
12 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
|
|
| 45 | c3o7kA_ |
|
not modelled |
26.3 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:ohcu decarboxylase;
PDBTitle: crystal structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline2 decarboxylase from klebsiella pneumoniae
|
|
|
| 46 | c3pq1A_ |
|
not modelled |
25.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:poly(a) rna polymerase;
PDBTitle: crystal structure of human mitochondrial poly(a) polymerase (papd1)
|
|
|
| 47 | c2a4aB_ |
|
not modelled |
25.2 |
19 |
PDB header:lyase
Chain: B: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: deoxyribose-phosphate aldolase from p. yoelii
|
|
|
| 48 | c5l3xB_ |
|
not modelled |
22.2 |
20 |
PDB header:transcription
Chain: B: PDB Molecule:negative elongation factor c/d;
PDBTitle: crystal structure of negative elongation factor subcomplex nelf-ac
|
|
|
| 49 | c2jq5A_ |
|
not modelled |
22.0 |
17 |
PDB header:structural genomics
Chain: A: PDB Molecule:sec-c motif;
PDBTitle: solution structure of rpa3114, a sec-c motif containing protein from2 rhodopseudomonas palustris; northeast structural genomics consortium3 target rpt5 / ontario center for structural proteomics target rp3097
|
|
|
| 50 | c5l9wB_ |
|
not modelled |
21.8 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 51 | c3ngjC_ |
|
not modelled |
21.8 |
20 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of a putative deoxyribose-phosphate aldolase from2 entamoeba histolytica
|
|
|
| 52 | d1vi7a1 |
|
not modelled |
20.2 |
35 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
|
|
| 53 | c4hadD_ |
|
not modelled |
19.7 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
|
|
|
| 54 | c3qyqC_ |
|
not modelled |
19.6 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:deoxyribose-phosphate aldolase, putative;
PDBTitle: 1.8 angstrom resolution crystal structure of a putative deoxyribose-2 phosphate aldolase from toxoplasma gondii me49
|
|
|
| 55 | c3hiyA_ |
|
not modelled |
19.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:minor editosome-associated tutase;
PDBTitle: minor editosome-associated tutase 1 with bound utp and mg
|
|
|
| 56 | c3ng3A_ |
|
not modelled |
19.3 |
37 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: crystal structure of deoxyribose phosphate aldolase from mycobacterium2 avium 104 in a schiff base with an unknown aldehyde
|
|
|
| 57 | c6e0oA_ |
|
not modelled |
19.2 |
16 |
PDB header:transferase/rna
Chain: A: PDB Molecule:cgas/dncv-like nucleotidyltransferase in e. coli homolog;
PDBTitle: structure of elizabethkingia meningoseptica cdne cyclic dinucleotide2 synthase with pppa[3'-5']pa
|
|
|
| 58 | c4mp6A_ |
|
not modelled |
18.5 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative ornithine cyclodeaminase;
PDBTitle: staphyloferrin b precursor biosynthetic enzyme sbnb bound to citrate2 and nad+
|
|
|
| 59 | d1h6da1 |
|
not modelled |
18.4 |
13 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 60 | d1jqra_ |
|
not modelled |
18.1 |
26 |
Fold:Nucleotidyltransferase
Superfamily:Nucleotidyltransferase
Family:DNA polymerase beta-like |
|
|
| 61 | c3pq1B_ |
|
not modelled |
17.9 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:poly(a) rna polymerase;
PDBTitle: crystal structure of human mitochondrial poly(a) polymerase (papd1)
|
|
|
| 62 | c5wu6B_ |
|
not modelled |
17.9 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:speckle targeted pip5k1a-regulated poly(a) polymerase;
PDBTitle: crystal structure of apo human tut1, form iv
|
|
|
| 63 | c5a2zB_ |
|
not modelled |
17.6 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:mitochondrial protein;
PDBTitle: crystal structure of mtpap in complex with gtp
|
|
|
| 64 | c6bqiA_ |
|
not modelled |
17.6 |
19 |
PDB header:translation
Chain: A: PDB Molecule:protein impact homolog;
PDBTitle: structure of two-domain translational regulator yih1 reveals a2 possible mechanism of action
|
|
|
| 65 | c3ceaA_ |
|
not modelled |
16.6 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
|
|
| 66 | d2cvea1 |
|
not modelled |
16.6 |
44 |
Fold:Ribosomal protein S5 domain 2-like
Superfamily:Ribosomal protein S5 domain 2-like
Family:YigZ N-terminal domain-like |
|
|
| 67 | d1f99a_ |
|
not modelled |
16.3 |
15 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
|
|
| 68 | c8iczA_ |
|
not modelled |
16.3 |
15 |
PDB header:transferase/dna
Chain: A: PDB Molecule:protein (dna polymerase beta (e.c.2.7.7.7));
PDBTitle: dna polymerase beta (pol b) (e.c.2.7.7.7) complexed with2 seven base pairs of dna; soaked in the presence of of datp3 (1 millimolar), mncl2 (5 millimolar), and lithium sulfate4 (75 millimolar)
|
|
|
| 69 | d2nvwa1 |
|
not modelled |
16.0 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 70 | c5uibA_ |
|
not modelled |
15.9 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase protein;
PDBTitle: crystal structure of an oxidoreductase from agrobacterium radiobacter2 in complex with nad+, l-tartaric acid and magnesium
|
|
|
| 71 | c3f2eA_ |
|
not modelled |
15.9 |
15 |
PDB header:viral protein
Chain: A: PDB Molecule:sirv coat protein;
PDBTitle: crystal structure of yellowstone sirv coat protein c-terminus
|
|
|
| 72 | c2glxD_ |
|
not modelled |
15.8 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:1,5-anhydro-d-fructose reductase;
PDBTitle: crystal structure analysis of bacterial 1,5-af reductase
|
|
|
| 73 | d1qmga2 |
|
not modelled |
15.1 |
42 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:6-phosphogluconate dehydrogenase-like, N-terminal domain |
|
|
| 74 | c3uuwB_ |
|
not modelled |
14.9 |
9 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase with nad(p)-binding rossmann-fold
PDBTitle: 1.63 angstrom resolution crystal structure of dehydrogenase (mvim)2 from clostridium difficile.
|
|
|
| 75 | c3nt5B_ |
|
not modelled |
14.8 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inositol 2-dehydrogenase/d-chiro-inositol 3-dehydrogenase;
PDBTitle: crystal structure of myo-inositol dehydrogenase from bacillus subtilis2 with bound cofactor and product inosose
|
|
|
| 76 | c6ajrA_ |
|
not modelled |
14.5 |
11 |
PDB header:dna binding protein
Chain: A: PDB Molecule:uracil dna glycosylase superfamily protein;
PDBTitle: complex form of uracil dna glycosylase x and uracil
|
|
|
| 77 | d1r8ka_ |
|
not modelled |
14.3 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 78 | c2q0dA_ |
|
not modelled |
13.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:rna uridylyl transferase;
PDBTitle: terminal uridylyl transferase 4 from trypanosoma brucei with bound atp
|
|
|
| 79 | d1p1xa_ |
|
not modelled |
13.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 80 | c5muuI_ |
|
not modelled |
13.6 |
17 |
PDB header:virus
Chain: I: PDB Molecule:major outer capsid protein;
PDBTitle: dsrna bacteriophage phi6 nucleocapsid
|
|
|
| 81 | c2ihmA_ |
|
not modelled |
13.6 |
14 |
PDB header:transferase/dna
Chain: A: PDB Molecule:dna polymerase mu;
PDBTitle: polymerase mu in ternary complex with gapped 11mer dna duplex and2 bound incoming nucleotide
|
|
|
| 82 | c4zbzA_ |
|
not modelled |
13.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:uracil-dna glycosylase;
PDBTitle: family 4 uracil-dna glycosylase from sulfolobus tokodaii (free form,2 x-ray wavelength=1.5418)
|
|
|
| 83 | d2fi0a1 |
|
not modelled |
13.4 |
13 |
Fold:SP0561-like
Superfamily:SP0561-like
Family:SP0561-like |
|
|
| 84 | c1evjC_ |
|
not modelled |
13.1 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: crystal structure of glucose-fructose oxidoreductase (gfor)2 delta1-22 s64d
|
|
|
| 85 | c4xbsA_ |
|
not modelled |
13.0 |
41 |
PDB header:lyase
Chain: A: PDB Molecule:deoxyribose-phosphate aldolase;
PDBTitle: 2-deoxyribose-5-phosphate aldolase mutant - e78k
|
|
|
| 86 | c2bn5A_ |
|
not modelled |
12.8 |
26 |
PDB header:nuclear protein
Chain: A: PDB Molecule:psi;
PDBTitle: p-element somatic inhibitor protein complex with u1-70k2 proline-rich peptide
|
|
|
| 87 | d1phna_ |
|
not modelled |
12.5 |
8 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
|
|
| 88 | d1jboa_ |
|
not modelled |
12.2 |
16 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
|
|
| 89 | c2n5xA_ |
|
not modelled |
12.2 |
50 |
PDB header:chaperone
Chain: A: PDB Molecule:hsp90 co-chaperone cdc37;
PDBTitle: c-terminal domain of cdc37 cochaperone
|
|
|
| 90 | c5wu5C_ |
|
not modelled |
12.1 |
19 |
PDB header:transferase
Chain: C: PDB Molecule:speckle targeted pip5k1a-regulated poly(a) polymerase;
PDBTitle: crystal structure of apo human tut1, form iii
|
|
|
| 91 | c2ho3D_ |
|
not modelled |
11.5 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, gfo/idh/moca family;
PDBTitle: crystal structure of oxidoreductase, gfo/idh/moca family from2 streptococcus pneumoniae
|
|
|
| 92 | d1cpca_ |
|
not modelled |
11.1 |
14 |
Fold:Globin-like
Superfamily:Globin-like
Family:Phycocyanin-like phycobilisome proteins |
|
|
| 93 | c3r66A_ |
|
not modelled |
11.0 |
13 |
PDB header:viral protein/antiviral protein
Chain: A: PDB Molecule:non-structural protein 1;
PDBTitle: crystal structure of human isg15 in complex with ns1 n-terminal region2 from influenza virus b, northeast structural genomics consortium3 target ids hx6481, hr2873, and or2
|
|
|
| 94 | d1xeqa1 |
|
not modelled |
10.9 |
13 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:N-terminal, RNA-binding domain of nonstructural protein NS1 |
|
|
| 95 | c5e4rA_ |
|
not modelled |
10.8 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ketol-acid reductoisomerase;
PDBTitle: crystal structure of domain-duplicated synthetic class ii ketol-acid2 reductoisomerase 2ia_kari-dd
|
|
|
| 96 | c2bn6A_ |
|
not modelled |
10.8 |
28 |
PDB header:nuclear protein
Chain: A: PDB Molecule:psi;
PDBTitle: p-element somatic inhibitor protein
|
|
|
| 97 | c5ncaA_ |
|
not modelled |
10.7 |
33 |
PDB header:structural protein
Chain: A: PDB Molecule:competence protein comgc;
PDBTitle: solution structure of comgc from streptococcus pneumoniae
|
|
|
| 98 | d1aora1 |
|
not modelled |
10.5 |
9 |
Fold:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Superfamily:Aldehyde ferredoxin oxidoreductase, C-terminal domains
Family:Aldehyde ferredoxin oxidoreductase, C-terminal domains |
|
|
| 99 | c3o3nA_ |
|
not modelled |
10.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:alpha-subunit 2-hydroxyisocaproyl-coa dehydratase;
PDBTitle: (r)-2-hydroxyisocaproyl-coa dehydratase in complex with its substrate2 (r)-2-hydroxyisocaproyl-coa
|
|
|