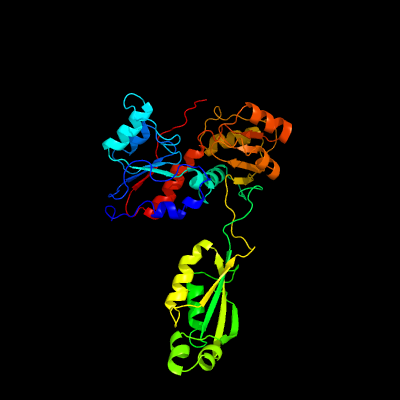
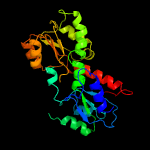
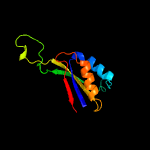
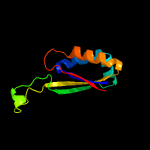
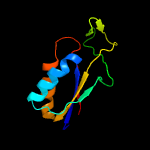
1 c2gx8B_
100.0
34
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: nif3-related protein;PDBTitle: the crystal structure of bacillus cereus protein related to nif3
2 d2gx8a1
100.0
34
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
3 c2nydB_
100.0
29
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
4 d2fywa1
100.0
27
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
5 d1nmpa_
100.0
26
Fold: NIF3 (NGG1p interacting factor 3)-likeSuperfamily: NIF3 (NGG1p interacting factor 3)-likeFamily: NIF3 (NGG1p interacting factor 3)-like
6 c4iwmD_
100.0
25
PDB header: unknown functionChain: D: PDB Molecule: upf0135 protein mj0927;PDBTitle: crystal structure of the conserved hypothetical protein mj0927 from2 methanocaldococcus jannaschii (in p21 form)
7 c2yybA_
100.0
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein ttha1606;PDBTitle: crystal structure of ttha1606 from thermus thermophilus hb8
8 c3rxyA_
99.1
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: nif3 protein;PDBTitle: crystal structure of nif3 superfamily protein from sphaerobacter2 thermophilus
9 c3mhyC_
95.3
19
PDB header: signaling proteinChain: C: PDB Molecule: pii-like protein pz;PDBTitle: a new pii protein structure
10 d1vfja_
94.9
23
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
11 c2rd5D_
94.7
22
PDB header: protein bindingChain: D: PDB Molecule: pii protein;PDBTitle: structural basis for the regulation of n-acetylglutamate kinase by pii2 in arabidopsis thaliana
12 c4usiC_
94.4
18
PDB header: signaling proteinChain: C: PDB Molecule: nitrogen regulatory protein pii;PDBTitle: nitrogen regulatory protein pii from chlamydomonas2 reinhardtii in complex with mgatp and 2-oxoglutarate
13 c3ahpA_
94.2
15
PDB header: electron transportChain: A: PDB Molecule: cuta1;PDBTitle: crystal structure of stable protein, cuta1, from a psychrotrophic2 bacterium shewanella sp. sib1
14 d2piia_
94.2
17
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
15 d2ns1b1
94.0
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
16 d1qy7a_
94.0
22
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
17 d1nzaa_
93.9
20
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
18 c4r25A_
93.9
19
PDB header: transcriptionChain: A: PDB Molecule: nitrogen regulatory pii-like protein;PDBTitle: structure of b. subtilis glnk
19 c4e98C_
93.9
18
PDB header: signaling proteinChain: C: PDB Molecule: cuta1 divalent ion tolerance protein;PDBTitle: crystal structure of possible cuta1 divalent ion tolerance protein2 from cryptosporidium parvum iowa ii
20 c2nuhA_
93.7
18
PDB header: unknown functionChain: A: PDB Molecule: periplasmic divalent cation tolerance protein;PDBTitle: crystal structure of cuta from the phytopathgen bacterium xylella2 fastidiosa
21 c4iyqB_
not modelled
93.7
14
PDB header: protein bindingChain: B: PDB Molecule: divalent ion tolerance protein cuta1;PDBTitle: crystal structure of divalent ion tolerance protein cuta1 from2 ehrlichia chaffeensis
22 c3o8wA_
not modelled
93.6
30
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen regulatory protein p-ii (glnb-1);PDBTitle: archaeoglobus fulgidus glnk1
23 d1ukua_
not modelled
93.5
18
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
24 d1vhfa_
not modelled
93.4
20
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
25 d1naqa_
not modelled
93.3
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
26 d1p1la_
not modelled
93.3
22
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
27 c2zomC_
not modelled
93.1
16
PDB header: unknown functionChain: C: PDB Molecule: protein cuta, chloroplast, putative, expressed;PDBTitle: crystal structure of cuta1 from oryza sativa
28 c6gdxA_
not modelled
93.1
16
PDB header: metal binding proteinChain: A: PDB Molecule: periplasmic divalent cation tolerance protein;PDBTitle: structure of cuta from synechococcus elongatus pcc7942 complexed with2 3 molecules of bis-tris
29 c2zfhA_
not modelled
93.0
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: cuta;PDBTitle: crystal structure of putative cuta1 from homo sapiens at 2.05a2 resolution
30 d1kr4a_
not modelled
92.7
20
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
31 c1xk8A_
not modelled
92.4
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: divalent cation tolerant protein cuta;PDBTitle: divalent cation tolerant protein cuta from homo sapiens o60888
32 d2zfha1
not modelled
92.3
16
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
33 c4ozlA_
not modelled
91.4
27
PDB header: signaling proteinChain: A: PDB Molecule: nitrogen regulatory protein p-ii;PDBTitle: glnk2 from haloferax mediterranei complexed with amp
34 d1hwua_
not modelled
90.9
22
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
35 c3ncpD_
not modelled
90.8
32
PDB header: signaling proteinChain: D: PDB Molecule: nitrogen regulatory protein p-ii (glnb-2);PDBTitle: glnk2 from archaeoglobus fulgidus
36 c3bzqA_
not modelled
90.4
23
PDB header: signaling protein/transcriptionChain: A: PDB Molecule: nitrogen regulatory protein p-ii;PDBTitle: high resolution crystal structure of nitrogen regulatory protein2 (rv2919c) of mycobacterium tuberculosis
37 c2j9dG_
not modelled
90.3
31
PDB header: membrane transportChain: G: PDB Molecule: hypothetical nitrogen regulatory pii-likePDBTitle: structure of glnk1 with bound effectors indicates2 regulatory mechanism for ammonia uptake
38 d1ul3a_
not modelled
89.1
22
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
39 c4s1nA_
not modelled
88.6
14
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: the crystal structure of phosphoribosylglycinamide formyltransferase2 from streptococcus pneumoniae tigr4
40 c3l7pA_
not modelled
87.4
35
PDB header: transcriptionChain: A: PDB Molecule: putative nitrogen regulatory protein pii;PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
41 d2cz4a1
not modelled
86.2
13
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Prokaryotic signal transducing protein
42 c3ce8A_
not modelled
82.6
13
PDB header: unknown functionChain: A: PDB Molecule: putative pii-like nitrogen regulatory protein;PDBTitle: crystal structure of a duf3240 family protein (sbal_0098) from2 shewanella baltica os155 at 2.40 a resolution
43 d1osce_
not modelled
81.7
12
Fold: Ferredoxin-likeSuperfamily: GlnB-likeFamily: Divalent ion tolerance proteins CutA (CutA1)
44 c5ghrA_
not modelled
79.6
14
PDB header: dna binding protein/replicationChain: A: PDB Molecule: ssdna-specific exonuclease;PDBTitle: dna replication protein
45 c3av3A_
not modelled
78.7
17
PDB header: transferaseChain: A: PDB Molecule: phosphoribosylglycinamide formyltransferase;PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 geobacillus kaustophilus
46 c2yx6C_
not modelled
76.3
12
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein ph0822;PDBTitle: crystal structure of ph0822
47 c5d4pA_
not modelled
72.7
15
PDB header: signaling proteinChain: A: PDB Molecule: putative nitrogen regulatory protein p-ii glnb;PDBTitle: structure of cpii bound to adp and bicarbonate, from thiomonas2 intermedia k12
48 c6cauA_
not modelled
71.4
15
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
49 c5f56A_
not modelled
70.7
22
PDB header: dna binding protein/dnaChain: A: PDB Molecule: single-stranded-dna-specific exonuclease;PDBTitle: structure of recj complexed with dna and ssb-ct
50 c3zu3A_
not modelled
69.4
9
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ypo4104/y4119/yp_4011;PDBTitle: structure of the enoyl-acp reductase fabv from yersinia pestis with2 the cofactor nadh (mr, cleaved histag)
51 d1p3da1
not modelled
69.3
16
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
52 c5ebbA_
not modelled
68.3
15
PDB header: hydrolaseChain: A: PDB Molecule: acid sphingomyelinase-like phosphodiesterase 3a;PDBTitle: structure of human sphingomyelinase phosphodiesterase like 3a2 (smpdl3a) with zn2+
53 c2ph5A_
not modelled
68.3
11
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
54 c5karA_
not modelled
67.8
15
PDB header: hydrolaseChain: A: PDB Molecule: acid sphingomyelinase-like phosphodiesterase 3b;PDBTitle: murine acid sphingomyelinase-like phosphodiesterase 3b (smpdl3b)
55 d2c1ha1
not modelled
66.5
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
56 c3k6jA_
not modelled
66.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: protein f01g10.3, confirmed by transcript evidence;PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
57 c4bubA_
not modelled
63.0
18
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate--ld-lysinePDBTitle: crystal structure of mure ligase from thermotoga maritima2 in complex with adp
58 c4euhA_
not modelled
62.3
9
PDB header: oxidoreductaseChain: A: PDB Molecule: putative reductase ca_c0462;PDBTitle: crystal structure of clostridium acetobutulicum trans-2-enoyl-coa2 reductase apo form
59 c5yh1A_
not modelled
60.3
17
PDB header: ribosomal proteinChain: A: PDB Molecule: member of s1p family of ribosomal proteins;PDBTitle: member of s1p family of ribosomal proteins pf0399 dhh domain
60 c2pv7B_
not modelled
60.1
15
PDB header: isomerase, oxidoreductaseChain: B: PDB Molecule: t-protein [includes: chorismate mutase (ec 5.4.99.5) (cm)PDBTitle: crystal structure of chorismate mutase / prephenate dehydrogenase2 (tyra) (1574749) from haemophilus influenzae rd at 2.00 a resolution
61 c3p19A_
not modelled
59.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: putative blue fluorescent protein;PDBTitle: improved nadph-dependent blue fluorescent protein
62 c3obkH_
not modelled
57.9
18
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
63 c3u49B_
not modelled
57.6
18
PDB header: oxidoreductaseChain: B: PDB Molecule: bacilysin biosynthesis oxidoreductase ywfh;PDBTitle: crystal structure of ywfh, nadph dependent reductase involved in2 bacilysin biosynthesis
64 d2p10a1
not modelled
55.5
22
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Mll9387-like
65 c5lzlH_
not modelled
54.3
17
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase
66 c5ldgA_
not modelled
54.0
25
PDB header: oxidoreductaseChain: A: PDB Molecule: (-)-isopiperitenone reductase;PDBTitle: isopiperitenone reductase from mentha piperita in complex with2 isopiperitenone and nadp
67 c2f00A_
not modelled
53.4
21
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate--l-alanine ligase;PDBTitle: escherichia coli murc
68 c5nmwA_
not modelled
53.1
33
PDB header: oxidoreductaseChain: A: PDB Molecule: flavin-containing monooxygenase;PDBTitle: crystal structure of the pyrrolizidine alkaloid n-oxygenase from2 zonocerus variegatus in complex with fad
69 c3nywD_
not modelled
52.2
21
PDB header: oxidoreductaseChain: D: PDB Molecule: putative oxidoreductase;PDBTitle: crystal structure of a betaketoacyl-[acp] reductase (fabg) from2 bacteroides thetaiotaomicron
70 c3v2hB_
not modelled
51.7
19
PDB header: oxidoreductaseChain: B: PDB Molecule: d-beta-hydroxybutyrate dehydrogenase;PDBTitle: the crystal structure of d-beta-hydroxybutyrate dehydrogenase from2 sinorhizobium meliloti
71 c5l53A_
not modelled
50.5
21
PDB header: oxidoreductaseChain: A: PDB Molecule: (-)-menthone:(+)-neomenthol reductase;PDBTitle: menthone neomenthol reductase from mentha piperita in complex with2 nadp
72 d1j6ua1
not modelled
50.3
15
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
73 d1vqta1
not modelled
50.3
15
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Decarboxylase
74 c3o26A_
not modelled
49.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: salutaridine reductase;PDBTitle: the structure of salutaridine reductase from papaver somniferum.
75 c5o3zK_
not modelled
49.3
32
PDB header: oxidoreductaseChain: K: PDB Molecule: sorbitol-6-phosphate dehydrogenase;PDBTitle: crystal structure of sorbitol-6-phosphate 2-dehydrogenase srld from2 erwinia amylovora
76 c4y96B_
not modelled
49.2
17
PDB header: isomeraseChain: B: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of triosephosphate isomerase from gemmata2 obscuriglobus
77 c2dp3A_
not modelled
48.9
21
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of a double mutant (c202a/a198v) of triosephosphate2 isomerase from giardia lamblia
78 d1n55a_
not modelled
48.7
17
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
79 c2nq8B_
not modelled
48.4
14
PDB header: oxidoreductaseChain: B: PDB Molecule: enoyl-acyl carrier reductase;PDBTitle: malarial enoyl acyl acp reductase bound with inh-nad adduct
80 c2foiB_
not modelled
48.4
14
PDB header: oxidoreductaseChain: B: PDB Molecule: enoyl-acyl carrier reductase;PDBTitle: synthesis, biological activity, and x-ray crystal structural analysis2 of diaryl ether inhibitors of malarial enoyl acp reductase.
81 c6bveA_
not modelled
48.2
19
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: triosephosphate isomerase of synechocystis in complex with 2-2 phosphoglycolic acid
82 d1p33a_
not modelled
48.0
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
83 c5ujwD_
not modelled
47.6
17
PDB header: isomeraseChain: D: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of triosephosphate isomerase from francisella2 tularensis subsp. tularensis schu s4
84 c2qhxB_
not modelled
46.2
21
PDB header: oxidoreductaseChain: B: PDB Molecule: pteridine reductase 1;PDBTitle: structure of pteridine reductase from leishmania major2 complexed with a ligand
85 c2p10D_
not modelled
45.9
22
PDB header: hydrolaseChain: D: PDB Molecule: mll9387 protein;PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
86 c5b1yB_
not modelled
45.7
11
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] reductase;PDBTitle: crystal structure of nadph bound carbonyl reductase from aeropyrum2 pernix
87 d1t3va_
not modelled
45.6
14
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
88 c4x54A_
not modelled
45.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase family;PDBTitle: crystal structure of an oxidoreductase (short chain2 dehydrogenase/reductase) from brucella ovis
89 c3t4xA_
not modelled
45.5
11
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase family;PDBTitle: short chain dehydrogenase/reductase family oxidoreductase from2 bacillus anthracis str. ames ancestor
90 c3s6dA_
not modelled
44.8
23
PDB header: isomeraseChain: A: PDB Molecule: putative triosephosphate isomerase;PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
91 d1ru8a_
not modelled
44.8
17
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: N-type ATP pyrophosphatases
92 c2wfbA_
not modelled
44.8
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative uncharacterized protein orp;PDBTitle: high resolution structure of the apo form of the orange2 protein (orp) from desulfovibrio gigas
93 d1m6ja_
not modelled
44.8
28
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
94 c2dknA_
not modelled
44.6
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 3-alpha-hydroxysteroid dehydrogenase;PDBTitle: crystal structure of the 3-alpha-hydroxysteroid dehydrogenase from2 pseudomonas sp. b-0831 complexed with nadh
95 d1uh5a_
not modelled
44.5
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
96 c4nbwA_
not modelled
44.3
25
PDB header: oxidoreductaseChain: A: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: crystal structure of fabg from plesiocystis pacifica
97 c6oz7A_
not modelled
44.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: uncharacterized oxidoreductase yohf;PDBTitle: putative oxidoreductase from escherichia coli str. k-12
98 d1eo1a_
not modelled
44.0
19
Fold: Ribonuclease H-like motifSuperfamily: Nitrogenase accessory factor-likeFamily: MTH1175-like
99 d1o5xa_
not modelled
43.6
21
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
100 c6m9uA_
not modelled
43.3
22
PDB header: oxidoreductaseChain: A: PDB Molecule: oxidoreductase, short chain dehydrogenase/reductase familyPDBTitle: structure of the apo-form of 20beta-hydroxysteroid dehydrogenase from2 bifidobacterium adolescentis strain l2-32
101 c6nrpC_
not modelled
43.2
21
PDB header: oxidoreductaseChain: C: PDB Molecule: 3-oxoacyl-acp reductase fabg;PDBTitle: putative short-chain dehydrogenase/reductase (sdr) from acinetobacter2 baumannii
102 c4nbrA_
not modelled
43.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical 3-oxoacyl-(acyl-carrier protein) reductase;PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier protein] reductase from2 brucella melitensis atcc 23457
103 c6j4nD_
not modelled
42.8
11
PDB header: hydrolaseChain: D: PDB Molecule: metallo-beta-lactamases pngm-1;PDBTitle: structure of papua new guinea mbl-1(pngm-1) native
104 c4obtA_
not modelled
42.7
19
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase, cytosolic;PDBTitle: crystal structure of arabidopsis thaliana cytosolic triose phosphate2 isomerase
105 c1j6uA_
not modelled
42.7
18
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate-alanine ligase murc;PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
106 d1kv5a_
not modelled
42.2
19
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
107 c5idxB_
not modelled
41.6
22
PDB header: oxidoreductaseChain: B: PDB Molecule: short-chain dehydrogenase/reductase sdr;PDBTitle: crystal structure of an oxidoreductase from burkholderia vietnamiensis
108 c6ixjK_
not modelled
41.5
21
PDB header: cytosolic proteinChain: K: PDB Molecule: sulfoacetaldehyde reductase;PDBTitle: the crystal structure of sulfoacetaldehyde reductase from klebsiella2 oxytoca
109 c4mknA_
not modelled
41.3
26
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase;PDBTitle: crystal structure of chloroplastic triosephosphate isomerase from2 chlamydomonas reinhardtii at 1.1 a of resolution
110 d1suxa_
not modelled
41.2
21
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
111 d1neya_
not modelled
41.2
17
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
112 c3s55F_
not modelled
40.2
19
PDB header: oxidoreductaseChain: F: PDB Molecule: putative short-chain dehydrogenase/reductase;PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from mycobacterium abscessus bound to nad
113 c2ntnB_
not modelled
39.9
22
PDB header: oxidoreductaseChain: B: PDB Molecule: 3-oxoacyl-[acyl-carrier-protein] reductase;PDBTitle: crystal structure of maba-c60v/g139a/s144l
114 c3qstA_
not modelled
39.8
13
PDB header: isomeraseChain: A: PDB Molecule: triosephosphate isomerase, putative;PDBTitle: crystal structure of trichomonas vaginalis triosephosphate isomerase2 tvag_096350 gene (val-45 variant)
115 d1b9ba_
not modelled
39.6
17
Fold: TIM beta/alpha-barrelSuperfamily: Triosephosphate isomerase (TIM)Family: Triosephosphate isomerase (TIM)
116 c1vjtA_
not modelled
39.5
10
PDB header: hydrolaseChain: A: PDB Molecule: alpha-glucosidase;PDBTitle: crystal structure of alpha-glucosidase (tm0752) from thermotoga2 maritima at 2.50 a resolution
117 d1sbya1
not modelled
39.1
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
118 c4yxfB_
not modelled
38.9
7
PDB header: oxidoreductaseChain: B: PDB Molecule: mups;PDBTitle: mups, a 3-oxoacyl (acp) reductase involved in mupirocin biosynthesis
119 c2wtzC_
not modelled
38.9
18
PDB header: ligaseChain: C: PDB Molecule: udp-n-acetylmuramoyl-l-alanyl-d-glutamate-PDBTitle: mure ligase of mycobacterium tuberculosis
120 c2p91A_
not modelled
38.7
15
PDB header: oxidoreductaseChain: A: PDB Molecule: enoyl-[acyl-carrier-protein] reductase [nadh];PDBTitle: crystal structure of enoyl-[acyl-carrier-protein] reductase (nadh)2 from aquifex aeolicus vf5