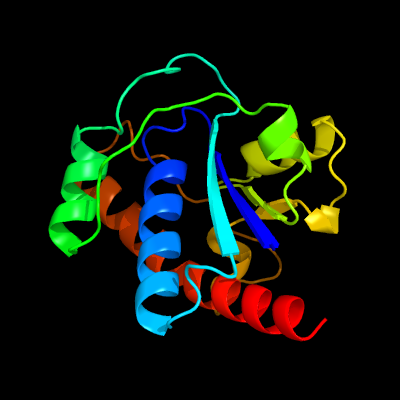
| 1 | c1u2pA_
|
|
 |
100.0 |
100 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: crystal structure of mycobacterium tuberculosis low2 molecular protein tyrosine phosphatase (mptpa) at 1.9a3 resolution
|
|
|
|
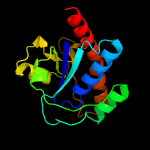
| 2 | c3jviA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein tyrosine phosphatase;
PDBTitle: product state mimic crystal structure of protein tyrosine phosphatase2 from entamoeba histolytica
|
|
|
|
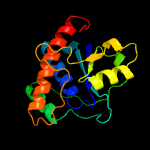
| 3 | d1dg9a_
|
|
 |
100.0 |
36 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
|
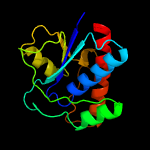
| 4 | d1d1qa_
|
|
 |
100.0 |
32 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
|
| 5 | d5pnta_
|
|
 |
100.0 |
33 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
|
| 6 | c2cwdA_
|
|
 |
100.0 |
45 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of tt1001 protein from thermus thermophilus hb8
|
|
|
|
| 7 | c2gi4A_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:possible phosphotyrosine protein phosphatase;
PDBTitle: solution structure of the low molecular weight protein2 tyrosine phosphatase from campylobacter jejuni.
|
|
|
|
| 8 | c5z3mB_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of low molecular weight phosphotyrosine phosphatase2 (vclmwptp-2) from vibrio choleraeo395
|
|
|
|
| 9 | c4etmB_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: B: PDB Molecule:low molecular weight protein-tyrosine-phosphatase yfkj;
PDBTitle: crystal structure of yfkj from bacillus subtilis
|
|
|
|
| 10 | c4lrqC_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: C: PDB Molecule:phosphotyrosine protein phosphatase;
PDBTitle: crystal structure of a low molecular weight phosphotyrosine2 phosphatase from vibrio choleraeo395
|
|
|
|
| 11 | c3rofA_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-phosphatase ptpa;
PDBTitle: crystal structure of the s. aureus protein tyrosine phosphatase ptpa
|
|
|
|
| 12 | c4etiA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-phosphatase ywle;
PDBTitle: crystal structure of ywle from bacillus subtilis
|
|
|
|
| 13 | c4picA_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:arginine phosphatase ywle;
PDBTitle: ywle arginine phosphatase from geobacillus stearothermophilus
|
|
|
|
| 14 | c1zggA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-
PDBTitle: solution structure of a low molecular weight protein2 tyrosine phosphatase from bacillus subtilis
|
|
|
|
| 15 | d1p8aa_
|
|
 |
100.0 |
29 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
|
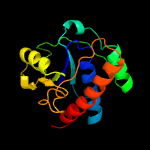
| 16 | c2fekA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight protein-tyrosine-
PDBTitle: structure of a protein tyrosine phosphatase
|
|
|
|
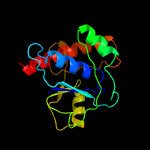
| 17 | c4d74A_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine-phosphatase amsi;
PDBTitle: 1.57 a crystal structure of erwinia amylovora tyrosine phosphatase2 amsi
|
|
|
|
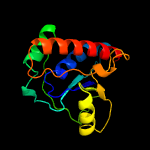
| 18 | c2wmyH_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: H: PDB Molecule:putative acid phosphatase wzb;
PDBTitle: crystal structure of the tyrosine phosphatase wzb from2 escherichia coli k30 in complex with sulphate.
|
|
|
|
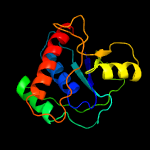
| 19 | c4egsB_
|
|
 |
100.0 |
31 |
PDB header:isomerase
Chain: B: PDB Molecule:ribose 5-phosphate isomerase rpib;
PDBTitle: crystal structure analysis of low molecular weight protein tyrosine2 phosphatase from t. tengcongensis
|
|
|
|
| 20 | c5gotA_
|
|
 |
100.0 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:low molecular weight phosphotyrosine phosphatase family
PDBTitle: crystal structure of sp-ptp, low molecular weight protein tyrosine2 phosphatase from streptococcus pyogenes
|
|
|
|
| 21 | d1jf8a_ |
|
not modelled |
100.0 |
26 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
| 22 | c2l18A_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: an arsenate reductase in the phosphate binding state
|
|
|
| 23 | c3rh0A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:arsenate reductase;
PDBTitle: corynebacterium glutamicum mycothiol/mycoredoxin1-dependent arsenate2 reductase cg_arsc2
|
|
|
| 24 | d1jl3a_ |
|
not modelled |
100.0 |
29 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
| 25 | d1y1la_ |
|
not modelled |
100.0 |
25 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:Phosphotyrosine protein phosphatases I
Family:Low-molecular-weight phosphotyrosine protein phosphatases |
|
|
| 26 | c5o7bA_ |
|
not modelled |
100.0 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative low molecular weight protein-tyrosine-phosphatase
PDBTitle: crystal structure of the slr0328 tyrosine phosphatase wzb from2 synechocystis sp. pcc 6803
|
|
|
| 27 | c3t38B_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:arsenate reductase;
PDBTitle: corynebacterium glutamicum thioredoxin-dependent arsenate reductase2 cg_arsc1'
|
|
|
| 28 | c3fdfA_ |
|
not modelled |
97.1 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:fr253;
PDBTitle: crystal structure of the serine phosphatase of rna2 polymerase ii ctd (ssu72 superfamily) from drosophila3 melanogaster. orthorhombic crystal form. northeast4 structural genomics consortium target fr253.
|
|
|
| 29 | c3o2sB_ |
|
not modelled |
96.6 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72 complex
|
|
|
| 30 | c3o2qB_ |
|
not modelled |
95.9 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:rna polymerase ii subunit a c-terminal domain phosphatase
PDBTitle: crystal structure of the human symplekin-ssu72-ctd phosphopeptide2 complex
|
|
|
| 31 | d1fmta2 |
|
not modelled |
93.7 |
22 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 32 | c5abrB_ |
|
not modelled |
90.8 |
34 |
PDB header:electron transport
Chain: B: PDB Molecule:ferredoxin, 2fe-2s;
PDBTitle: structure of fesi protein from azotobacter vinelandii
|
|
|
| 33 | c1tvmA_ |
|
not modelled |
90.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
|
|
| 34 | c1vkrA_ |
|
not modelled |
88.8 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
|
|
| 35 | d1vkra_ |
|
not modelled |
88.8 |
19 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
|
|
| 36 | c3czcA_ |
|
not modelled |
87.9 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from streptococcus2 mutans
|
|
|
| 37 | c1fmtA_ |
|
not modelled |
86.6 |
22 |
PDB header:formyltransferase
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
|
|
|
| 38 | c4mgeB_ |
|
not modelled |
85.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:pts system, cellobiose-specific iib component;
PDBTitle: 1.85 angstrom resolution crystal structure of pts system cellobiose-2 specific transporter subunit iib from bacillus anthracis.
|
|
|
| 39 | c3wfjD_ |
|
not modelled |
84.6 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: the complex structure of d-mandelate dehydrogenase with nadh
|
|
|
| 40 | c3nbmA_ |
|
not modelled |
84.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, lactose-specific iibc components;
PDBTitle: the lactose-specific iib component domain structure of the2 phosphoenolpyruvate:carbohydrate phosphotransferase system (pts) from3 streptococcus pneumoniae.
|
|
|
| 41 | c5gqsA_ |
|
not modelled |
80.0 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:pts galactitol transporter subunit iib;
PDBTitle: nmr based solution structure of pts system, galactitol-specific iib2 component from methicillin resistant staphylococcus aureus
|
|
|
| 42 | c2l2qA_ |
|
not modelled |
76.4 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, cellobiose-specific iib component (cela);
PDBTitle: solution structure of cellobiose-specific phosphotransferase iib2 component protein from borrelia burgdorferi
|
|
|
| 43 | c2iyaB_ |
|
not modelled |
74.3 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a blueprint2 for antibiotic engineering
|
|
|
| 44 | c4leiB_ |
|
not modelled |
71.3 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:ndp-forosamyltransferase;
PDBTitle: spinosyn forosaminyltransferase spnp
|
|
|
| 45 | c3eagA_ |
|
not modelled |
71.2 |
19 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate:l-alanyl-gamma-d-glutamyl-meso-
PDBTitle: the crystal structure of udp-n-acetylmuramate:l-alanyl-gamma-d-2 glutamyl-meso-diaminopimelate ligase (mpl) from neisseria3 meningitides
|
|
|
| 46 | d1f37b_ |
|
not modelled |
69.0 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
|
|
| 47 | c6norB_ |
|
not modelled |
67.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nad dependent dehydrogenase;
PDBTitle: crystal structure of gend2 from gentamicin a biosynthesis in complex2 with nad
|
|
|
| 48 | c6cauA_ |
|
not modelled |
64.9 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: udp-n-acetylmuramate--alanine ligase from acinetobacter baumannii2 ab5075-uw with amppnp
|
|
|
| 49 | d1ydga_ |
|
not modelled |
64.1 |
16 |
Fold:Flavodoxin-like
Superfamily:Flavoproteins
Family:WrbA-like |
|
|
| 50 | d2fug21 |
|
not modelled |
63.7 |
22 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:NQO2-like |
|
|
| 51 | c5zbyA_ |
|
not modelled |
62.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:hydrogenase maturation protease hyci;
PDBTitle: crystal structure of a [nife] hydrogenase maturation protease hyci2 from thermococcus kodakarensis kod1
|
|
|
| 52 | c2f00A_ |
|
not modelled |
62.1 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate--l-alanine ligase;
PDBTitle: escherichia coli murc
|
|
|
| 53 | c3beoA_ |
|
not modelled |
61.6 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:udp-n-acetylglucosamine 2-epimerase;
PDBTitle: a structural basis for the allosteric regulation of non-2 hydrolyzing udp-glcnac 2-epimerases
|
|
|
| 54 | c3rbvA_ |
|
not modelled |
61.6 |
22 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:sugar 3-ketoreductase;
PDBTitle: crystal structure of kijd10, a 3-ketoreductase from actinomadura2 kijaniata incomplex with nadp
|
|
|
| 55 | d1cfza_ |
|
not modelled |
57.2 |
20 |
Fold:Phosphorylase/hydrolase-like
Superfamily:HybD-like
Family:Hydrogenase maturating endopeptidase HybD |
|
|
| 56 | c3kcqA_ |
|
not modelled |
57.2 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: crystal structure of phosphoribosylglycinamide formyltransferase from2 anaplasma phagocytophilum
|
|
|
| 57 | d1u6ka1 |
|
not modelled |
54.9 |
16 |
Fold:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Superfamily:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Family:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) |
|
|
| 58 | c1yj8C_ |
|
not modelled |
54.6 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: initial structural analysis of plasmodium falciparum glycerol-3-2 phosphate dehydrogenase
|
|
|
| 59 | c2bibA_ |
|
not modelled |
53.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:teichoic acid phosphorylcholine esterase/ choline binding
PDBTitle: crystal structure of the complete modular teichioic acid2 phosphorylcholine esterase pce (cbpe) from streptococcus pneumoniae
|
|
|
| 60 | c5lc5E_ |
|
not modelled |
53.8 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nadh dehydrogenase [ubiquinone] flavoprotein 2,
PDBTitle: structure of mammalian respiratory complex i, class2
|
|
|
| 61 | c1j6uA_ |
|
not modelled |
53.5 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramate-alanine ligase murc;
PDBTitle: crystal structure of udp-n-acetylmuramate-alanine ligase murc (tm0231)2 from thermotoga maritima at 2.3 a resolution
|
|
|
| 62 | c5xt2C_ |
|
not modelled |
52.5 |
8 |
PDB header:dna binding protein
Chain: C: PDB Molecule:response regulator fixj;
PDBTitle: crystal structures of full-length fixj from b. japonicum crystallized2 in space group p212121
|
|
|
| 63 | d1iiba_ |
|
not modelled |
52.4 |
8 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
|
|
| 64 | d1vlva2 |
|
not modelled |
51.4 |
18 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 65 | d1zh8a1 |
|
not modelled |
48.6 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
| 66 | c3tqrA_ |
|
not modelled |
48.2 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoribosylglycinamide formyltransferase;
PDBTitle: structure of the phosphoribosylglycinamide formyltransferase (purn) in2 complex with ches from coxiella burnetii
|
|
|
| 67 | c1i36A_ |
|
not modelled |
48.1 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved hypothetical protein mth1747;
PDBTitle: structure of conserved protein mth1747 of unknown function reveals2 structural similarity with 3-hydroxyacid dehydrogenases
|
|
|
| 68 | c3ceaA_ |
|
not modelled |
45.5 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:myo-inositol 2-dehydrogenase;
PDBTitle: crystal structure of myo-inositol 2-dehydrogenase (np_786804.1) from2 lactobacillus plantarum at 2.40 a resolution
|
|
|
| 69 | c6gcsH_ |
|
not modelled |
45.2 |
10 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:24-kda subunit (nuhm);
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
|
|
|
| 70 | c3f4aA_ |
|
not modelled |
43.2 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein ygr203w;
PDBTitle: structure of ygr203w, a yeast protein tyrosine phosphatase of the2 rhodanese family
|
|
|
| 71 | c5b3uB_ |
|
not modelled |
42.3 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:biliverdin reductase;
PDBTitle: crystal structure of biliverdin reductase in complex with nadp+ from2 synechocystis sp. pcc 6803
|
|
|
| 72 | c2p6pB_ |
|
not modelled |
38.9 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:glycosyl transferase;
PDBTitle: x-ray crystal structure of c-c bond-forming dtdp-d-olivose-transferase2 urdgt2
|
|
|
| 73 | c3rfoA_ |
|
not modelled |
38.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:methionyl-trna formyltransferase;
PDBTitle: crystal structure of methyionyl-trna formyltransferase from bacillus2 anthracis
|
|
|
| 74 | c4f67A_ |
|
not modelled |
37.1 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0176 protein lpg2838;
PDBTitle: three dimensional structure of the double mutant of upf0176 protein2 lpg2838 from legionella pneumophila at the resolution 1.8a, northeast3 structural genomics consortium (nesg) target lgr82
|
|
|
| 75 | c4hadD_ |
|
not modelled |
36.8 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of probable oxidoreductase protein from rhizobium2 etli cfn 42
|
|
|
| 76 | c2auvA_ |
|
not modelled |
36.7 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:potential nad-reducing hydrogenase subunit;
PDBTitle: solution structure of hndac : a thioredoxin-like [2fe-2s]2 ferredoxin involved in the nadp-reducing hydrogenase3 complex
|
|
|
| 77 | d1duvg2 |
|
not modelled |
36.4 |
10 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 78 | c6hl2C_ |
|
not modelled |
36.3 |
16 |
PDB header:electron transport
Chain: C: PDB Molecule:nadh-quinone oxidoreductase subunit e;
PDBTitle: wild-type nuoef from aquifex aeolicus - oxidized form
|
|
|
| 79 | d1m2da_ |
|
not modelled |
35.4 |
23 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioredoxin-like 2Fe-2S ferredoxin |
|
|
| 80 | d1zaka1 |
|
not modelled |
35.1 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
|
|
| 81 | c3db2C_ |
|
not modelled |
34.8 |
13 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative nadph-dependent oxidoreductase;
PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
|
|
|
| 82 | c4ambB_ |
|
not modelled |
34.2 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:snogd;
PDBTitle: crystal structure of the glycosyltransferase snogd from streptomyces2 nogalater
|
|
|
| 83 | d1j6ua1 |
|
not modelled |
33.8 |
27 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 84 | c3aufA_ |
|
not modelled |
33.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glycinamide ribonucleotide transformylase 1;
PDBTitle: crystal structure of glycinamide ribonucleotide transformylase 1 from2 symbiobacterium toebii
|
|
|
| 85 | c3tsaA_ |
|
not modelled |
33.5 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:ndp-rhamnosyltransferase;
PDBTitle: spinosyn rhamnosyltransferase spng
|
|
|
| 86 | c2eq8C_ |
|
not modelled |
33.2 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:pyruvate dehydrogenase complex, dihydrolipoamide
PDBTitle: crystal structure of lipoamide dehydrogenase from thermus thermophilus2 hb8 with psbdp
|
|
|
| 87 | c4ehxA_ |
|
not modelled |
32.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:tetraacyldisaccharide 4'-kinase;
PDBTitle: crystal structure of lpxk from aquifex aeolicus at 1.9 angstrom2 resolution
|
|
|
| 88 | c2e85B_ |
|
not modelled |
32.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase 3 maturation protease;
PDBTitle: crystal structure of the hydrogenase 3 maturation protease
|
|
|
| 89 | c3kklA_ |
|
not modelled |
32.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable chaperone protein hsp33;
PDBTitle: crystal structure of functionally unknown hsp33 from2 saccharomyces cerevisiae
|
|
|
| 90 | c3uuwB_ |
|
not modelled |
31.4 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase with nad(p)-binding rossmann-fold
PDBTitle: 1.63 angstrom resolution crystal structure of dehydrogenase (mvim)2 from clostridium difficile.
|
|
|
| 91 | c5ijaB_ |
|
not modelled |
30.2 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:hydrogenase-specific maturation endopeptidase;
PDBTitle: [nife] hydrogenase maturation protease hybd from thermococcus2 kodakarensis
|
|
|
| 92 | c4qxzA_ |
|
not modelled |
29.9 |
18 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a hypothetical protein from staphylococcus aureus
|
|
|
| 93 | c3i2vA_ |
|
not modelled |
29.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:adenylyltransferase and sulfurtransferase mocs3;
PDBTitle: crystal structure of human mocs3 rhodanese-like domain
|
|
|
| 94 | c4atyA_ |
|
not modelled |
29.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:terephthalate 1,2-cis-dihydrodiol dehydrogenase;
PDBTitle: crystal structure of a terephthalate 1,2-cis-2 dihydrodioldehydrogenase from burkholderia xenovorans3 lb400
|
|
|
| 95 | c1zakB_ |
|
not modelled |
29.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:adenylate kinase;
PDBTitle: adenylate kinase from maize in complex with the inhibitor2 p1,p5-bis(adenosine-5'-)pentaphosphate (ap5a)
|
|
|
| 96 | c5vytD_ |
|
not modelled |
29.1 |
21 |
PDB header:transferase
Chain: D: PDB Molecule:gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-
PDBTitle: crystal structure of the wbkc n-formyltransferase (f142a variant) from2 brucella melitensis
|
|
|
| 97 | c1nm3B_ |
|
not modelled |
28.9 |
14 |
PDB header:electron transport
Chain: B: PDB Molecule:protein hi0572;
PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
|
|
|
| 98 | c2q62A_ |
|
not modelled |
28.7 |
11 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
|
|
| 99 | c3fojA_ |
|
not modelled |
28.6 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of ssp1007 from staphylococcus2 saprophyticus subsp. saprophyticus. northeast structural3 genomics target syr101a.
|
|
|
| 100 | c4mkzA_ |
|
not modelled |
28.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inositol dehydrogenase;
PDBTitle: crystal structure of apo scyllo-inositol dehydrogenase from2 lactobacillus casei at 77k
|
|
|
| 101 | c2uzqE_ |
|
not modelled |
27.4 |
14 |
PDB header:hydrolase
Chain: E: PDB Molecule:m-phase inducer phosphatase 2;
PDBTitle: protein phosphatase, new crystal form
|
|
|
| 102 | c1ofgF_ |
|
not modelled |
26.3 |
17 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:glucose-fructose oxidoreductase;
PDBTitle: glucose-fructose oxidoreductase
|
|
|
| 103 | c1h6dL_ |
|
not modelled |
26.3 |
17 |
PDB header:protein translocation
Chain: L: PDB Molecule:precursor form of glucose-fructose
PDBTitle: oxidized precursor form of glucose-fructose oxidoreductase2 from zymomonas mobilis complexed with glycerol
|
|
|
| 104 | c4fgwA_ |
|
not modelled |
25.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase [nad(+)] 1;
PDBTitle: structure of glycerol-3-phosphate dehydrogenase, gpd1, from2 sacharomyces cerevisiae
|
|
|
| 105 | d1r8ka_ |
|
not modelled |
25.7 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:PdxA-like |
|
|
| 106 | c2k0zA_ |
|
not modelled |
25.3 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein hp1203;
PDBTitle: solution nmr structure of protein hp1203 from helicobacter pylori2 26695. northeast structural genomics consortium (nesg) target3 pt1/ontario center for structural proteomics target hp1203
|
|
|
| 107 | c6gnfC_ |
|
not modelled |
24.8 |
21 |
PDB header:transferase
Chain: C: PDB Molecule:glycogen synthase;
PDBTitle: granule bound starch synthase from cyanobacterium sp. clg1 bound to2 acarbose and adp
|
|
|
| 108 | c3wadA_ |
|
not modelled |
24.6 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyltransferase;
PDBTitle: crystal structure of glycosyltransferase vinc involved in the2 biosynthesis of vicenistatin
|
|
|
| 109 | c2j6pF_ |
|
not modelled |
24.3 |
23 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:sb(v)-as(v) reductase;
PDBTitle: structure of as-sb reductase from leishmania major
|
|
|
| 110 | c3ktdC_ |
|
not modelled |
24.1 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:prephenate dehydrogenase;
PDBTitle: crystal structure of a putative prephenate dehydrogenase (cgl0226)2 from corynebacterium glutamicum atcc 13032 at 2.60 a resolution
|
|
|
| 111 | c3r0jA_ |
|
not modelled |
23.8 |
20 |
PDB header:dna binding protein
Chain: A: PDB Molecule:possible two component system response transcriptional
PDBTitle: structure of phop from mycobacterium tuberculosis
|
|
|
| 112 | d1p3da1 |
|
not modelled |
23.3 |
20 |
Fold:MurCD N-terminal domain
Superfamily:MurCD N-terminal domain
Family:MurCD N-terminal domain |
|
|
| 113 | d1dxha2 |
|
not modelled |
22.9 |
14 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
|
|
| 114 | c3ilmD_ |
|
not modelled |
22.8 |
13 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:alr3790 protein;
PDBTitle: crystal structure of the alr3790 protein from anabaena sp. northeast2 structural genomics consortium target nsr437h
|
|
|
| 115 | c4e12A_ |
|
not modelled |
22.7 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:diketoreductase;
PDBTitle: substrate-directed dual catalysis of dicarbonyl compounds by2 diketoreductase
|
|
|
| 116 | d1w85i_ |
|
not modelled |
22.3 |
15 |
Fold:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Superfamily:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex
Family:Peripheral subunit-binding domain of 2-oxo acid dehydrogenase complex |
|
|
| 117 | c3dv0I_ |
|
not modelled |
22.3 |
15 |
PDB header:oxidoreductase/transferase
Chain: I: PDB Molecule:dihydrolipoyllysine-residue acetyltransferase
PDBTitle: snapshots of catalysis in the e1 subunit of the pyruvate2 dehydrogenase multi-enzyme complex
|
|
|
| 118 | d2cq2a1 |
|
not modelled |
22.2 |
16 |
Fold:Ferredoxin-like
Superfamily:RNA-binding domain, RBD
Family:Canonical RBD |
|
|
| 119 | c5ayvB_ |
|
not modelled |
22.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of archaeal ketopantoate reductase complexed with2 coenzyme a and 2-oxopantoate
|
|
|
| 120 | c5zikC_ |
|
not modelled |
21.6 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:probable 2-dehydropantoate 2-reductase;
PDBTitle: crystal structure of ketopantoate reductase from pseudomonas2 aeruginosa
|
|
|