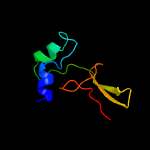
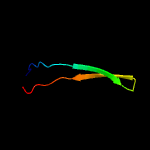
1 c2mc7A_
50.8
33
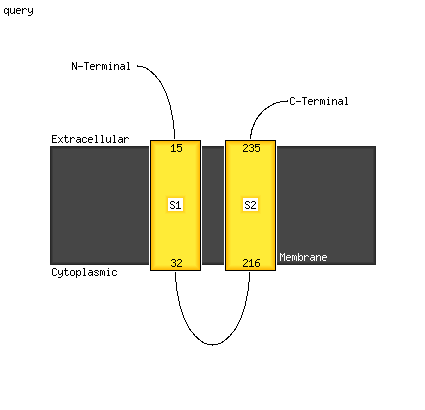
PDB header: membrane proteinChain: A: PDB Molecule: regulatory peptide;PDBTitle: structure of salmonella mgtr
2 c1zzaA_
33.4
12
PDB header: membrane proteinChain: A: PDB Molecule: stannin;PDBTitle: solution nmr structure of the membrane protein stannin
3 c5n9yB_
29.2
14
PDB header: membrane proteinChain: B: PDB Molecule: zinc transport protein zntb;PDBTitle: the full-length structure of zntb
4 c2lx0A_
28.6
14
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p14;PDBTitle: arced helix (arch) nmr structure of the reovirus p14 fusion-associated2 small transmembrane (fast) protein transmembrane domain (tmd) in3 dodecyl phosphocholine (dpc) micelles
5 c3tqzA_
28.0
17
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut)2 from coxiella burnetii
6 c6gcsW_
26.3
12
PDB header: oxidoreductaseChain: W: PDB Molecule: nb6m subunit;PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
7 d1sixa_
26.1
22
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
8 c2i5nH_
25.0
12
PDB header: photosynthesisChain: H: PDB Molecule: reaction center protein h chain;PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
9 c2bbjB_
23.4
24
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
10 c3ca9A_
20.2
19
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
11 d2r6ia1
18.3
15
Fold: ATP12-likeSuperfamily: ATP12-likeFamily: ATP12-like
12 c5lnkq_
18.2
17
PDB header: oxidoreductaseChain: Q: PDB Molecule: PDBTitle: entire ovine respiratory complex i
13 c5o31Z_
16.7
17
PDB header: oxidoreductaseChain: Z: PDB Molecule: nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunitPDBTitle: mitochondrial complex i in the deactive state
14 c6rdr8_
16.4
21
PDB header: proton transportChain: 8: PDB Molecule: mitochondrial atp synthase subunit asa8;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1d,2 monomer-masked refinement
15 c6rdi8_
16.4
21
PDB header: proton transportChain: 8: PDB Molecule: mitochondrial atp synthase subunit asa8;PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 1a,2 monomer-masked refinement
16 d1thqa_
15.4
33
Fold: Transmembrane beta-barrelsSuperfamily: OMPA-likeFamily: Outer membrane enzyme PagP
17 c6maiA_
15.3
19
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5'-triphosphate nucleotidohydrolase2 from legionella pneumophila philadelphia 1
18 c6mplA_
14.4
35
PDB header: membrane proteinChain: A: PDB Molecule: matrix protein 2;PDBTitle: racemic m2-tm i39a crystallized from racemic detergent
19 c2na6A_
14.2
26
PDB header: apoptosisChain: A: PDB Molecule: tumor necrosis factor receptor superfamily member 6;PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
20 c2na6B_
14.2
26
PDB header: apoptosisChain: B: PDB Molecule: tumor necrosis factor receptor superfamily member 6;PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
21 c2na6C_
not modelled
14.2
26
PDB header: apoptosisChain: C: PDB Molecule: tumor necrosis factor receptor superfamily member 6;PDBTitle: transmembrane domain of mouse fas/cd95 death receptor
22 c4xtnJ_
not modelled
14.1
6
PDB header: membrane proteinChain: J: PDB Molecule: sodium pumping rhodopsin;PDBTitle: crystal structure of the light-driven sodium pump kr2 in the2 pentameric red form, ph 4.9
23 d1xmeb2
not modelled
13.1
18
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region
24 c6roiC_
not modelled
12.8
17
PDB header: lipid transportChain: C: PDB Molecule: cell division control protein 50;PDBTitle: cryo-em structure of the partially activated drs2p-cdc50p
25 c3so2A_
not modelled
12.7
17
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: chlorella dutpase
26 c2voyE_
not modelled
12.7
18
PDB header: hydrolaseChain: E: PDB Molecule: sarcoplasmic/endoplasmic reticulum calcium atpase 1;PDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
27 c2p9oB_
not modelled
12.4
8
PDB header: hydrolaseChain: B: PDB Molecule: dutp pyrophosphatase-like protein;PDBTitle: structure of dutpase from arabidopsis thaliana
28 d2vvpa1
not modelled
12.0
17
Fold: Ribose/Galactose isomerase RpiB/AlsBSuperfamily: Ribose/Galactose isomerase RpiB/AlsBFamily: Ribose/Galactose isomerase RpiB/AlsB
29 c3ddlB_
not modelled
11.6
15
PDB header: transport proteinChain: B: PDB Molecule: xanthorhodopsin;PDBTitle: crystallographic structure of xanthorhodopsin, a light-driven ion pump2 with dual chromophore
30 d1sjna_
not modelled
11.2
22
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
31 c5xywD_
not modelled
9.9
67
PDB header: protein bindingChain: D: PDB Molecule: gd21652;PDBTitle: crystal structure of drosophila simulans rhino chromoshadow domain in2 complex with n-terminal domain
32 c4ev6E_
not modelled
9.8
7
PDB header: metal transportChain: E: PDB Molecule: magnesium transport protein cora;PDBTitle: the complete structure of cora magnesium transporter from2 methanocaldococcus jannaschii
33 d2iuba2
not modelled
9.7
25
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region
34 c3h6xA_
not modelled
9.7
19
PDB header: hydrolaseChain: A: PDB Molecule: dutpase;PDBTitle: crystal structure of dutpase from streptococcus mutans
35 c2kncA_
not modelled
9.5
38
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
36 c3c3iA_
not modelled
9.5
19
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
37 c6elhA_
not modelled
9.4
13
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide reductase;PDBTitle: low resolution structure of neisseria meningitidis qnor
38 c6mpnB_
not modelled
9.4
30
PDB header: membrane proteinChain: B: PDB Molecule: matrix protein 2;PDBTitle: racemic m2-tm i42e crystallized from racemic detergent
39 c6mpnA_
not modelled
9.4
30
PDB header: membrane proteinChain: A: PDB Molecule: matrix protein 2;PDBTitle: racemic m2-tm i42e crystallized from racemic detergent
40 c5kk2E_
not modelled
9.1
21
PDB header: membrane protein, transport protein, sigChain: E: PDB Molecule: voltage-dependent calcium channel gamma-2 subunit;PDBTitle: architecture of fully occupied glua2 ampa receptor - tarp complex2 elucidated by single particle cryo-electron microscopy
41 c3krbB_
not modelled
9.0
13
PDB header: oxidoreductaseChain: B: PDB Molecule: aldose reductase;PDBTitle: structure of aldose reductase from giardia lamblia at 1.75a resolution
42 c6adqP_
not modelled
9.0
20
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
43 c2axtc_
not modelled
8.7
14
PDB header: electron transportChain: C: PDB Molecule: photosystem ii cp43 protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
44 d2axtc1
not modelled
8.7
14
Fold: Photosystem II antenna protein-likeSuperfamily: Photosystem II antenna protein-likeFamily: Photosystem II antenna protein-like
45 d1ehkb2
not modelled
8.6
18
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region
46 c3sokB_
not modelled
8.3
11
PDB header: cell adhesionChain: B: PDB Molecule: fimbrial protein;PDBTitle: dichelobacter nodosus pilin fima
47 c4g0hA_
not modelled
7.9
20
PDB header: toxinChain: A: PDB Molecule: cytotoxicity-associated immunodominant antigen;PDBTitle: crystal structure of the n-terminal domain of helicobacter pylori caga2 protein
48 d1vmha_
not modelled
7.0
60
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
49 d1vpha_
not modelled
7.0
40
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
50 c5xyvC_
not modelled
7.0
50
PDB header: protein bindingChain: C: PDB Molecule: protein deadlock;PDBTitle: crystal structure of drosophila melanogaster rhino chromoshadow domain2 in complex with deadlock n-terminal domain
51 d1vmfa_
not modelled
6.9
60
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
52 c3lqwA_
not modelled
6.8
14
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5-triphosphate2 nucleotidohydrolase from entamoeba histolytica
53 c1fooA_
not modelled
6.8
17
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase;PDBTitle: bovine endothelial nitric oxide synthase heme domain complexed with l-2 arg and no(h4b-free)
54 d1nosa_
not modelled
6.6
15
Fold: Nitric oxide (NO) synthase oxygenase domainSuperfamily: Nitric oxide (NO) synthase oxygenase domainFamily: Nitric oxide (NO) synthase oxygenase domain
55 c1ve0A_
not modelled
6.6
60
PDB header: metal binding proteinChain: A: PDB Molecule: hypothetical protein (st2072);PDBTitle: crystal structure of uncharacterized protein st2072 from sulfolobus2 tokodaii
56 c2cu5C_
not modelled
6.6
60
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved hypothetical protein tt1486;PDBTitle: crystal structure of the conserved hypothetical protein tt1486 from2 thermus thermophilus hb8
57 d1vmja_
not modelled
6.5
60
Fold: YjbQ-likeSuperfamily: YjbQ-likeFamily: YjbQ-like
58 c2n1pA_
not modelled
6.5
21
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 5b, ns5b;PDBTitle: structure of the c-terminal membrane domain of hcv ns5b protein
59 c4xtrG_
not modelled
6.5
32
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: pep12p;PDBTitle: structure of get3 bound to the transmembrane domain of pep12
60 c3zf6A_
not modelled
6.4
9
PDB header: hydrolaseChain: A: PDB Molecule: dutpase;PDBTitle: phage dutpases control transfer of virulence genes by a proto-2 oncogenic g protein-like mechanism. (staphylococcus bacteriophage3 80alpha dutpase d81a d110c s168c mutant with dupnhpp).
61 c4rp9A_
not modelled
6.3
22
PDB header: membrane proteinChain: A: PDB Molecule: ascorbate-specific permease iic component ulaa;PDBTitle: bacterial vitamin c transporter ulaa/sgat in c2 form
62 d1om4a_
not modelled
6.2
20
Fold: Nitric oxide (NO) synthase oxygenase domainSuperfamily: Nitric oxide (NO) synthase oxygenase domainFamily: Nitric oxide (NO) synthase oxygenase domain
63 c2p6hB_
not modelled
6.1
60
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of hypothetical protein ape1520 from aeropyrum2 pernix k1
64 d1eysh2
not modelled
6.1
28
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction centre subunit H, transmembrane regionFamily: Photosystem II reaction centre subunit H, transmembrane region
65 c4kkpA_
not modelled
6.1
25
PDB header: structural proteinChain: A: PDB Molecule: rbma protein;PDBTitle: crystal structure of vibrio cholerae rbma (crystal form 2)
66 c4djiA_
not modelled
6.1
14
PDB header: transport proteinChain: A: PDB Molecule: probable glutamate/gamma-aminobutyrate antiporter;PDBTitle: structure of glutamate-gaba antiporter gadc
67 c2p6cB_
not modelled
6.0
80
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: aq_2013 protein;PDBTitle: crystal structure of hypothetical protein aq_2013 from aquifex2 aeolicus vf5.
68 c5azdA_
not modelled
5.9
16
PDB header: transport proteinChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: crystal structure of thermophilic rhodopsin.
69 d1euwa_
not modelled
5.8
19
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
70 c2okdB_
not modelled
5.7
11
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: high resolution crystal structures of vaccinia virus dutpase
71 c3ecyA_
not modelled
5.6
14
PDB header: hydrolaseChain: A: PDB Molecule: cg4584-pa, isoform a (bcdna.ld08534);PDBTitle: crystal structural analysis of drosophila melanogaster dutpase
72 c5zdnA_
not modelled
5.6
18
PDB header: hydrolaseChain: A: PDB Molecule: fomd;PDBTitle: the complex structure of fomd with cdp
73 c3am6C_
not modelled
5.6
22
PDB header: transport proteinChain: C: PDB Molecule: rhodopsin-2;PDBTitle: crystal structure of the proton pumping rhodopsin ar2 from marine alga2 acetabularia acetabulum
74 d1c6va_
not modelled
5.6
43
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Retroviral integrase, catalytic domain
75 c2an7A_
not modelled
5.5
17
PDB header: dna binding proteinChain: A: PDB Molecule: protein pard;PDBTitle: solution structure of the bacterial antidote pard
76 d1m9ma_
not modelled
5.2
17
Fold: Nitric oxide (NO) synthase oxygenase domainSuperfamily: Nitric oxide (NO) synthase oxygenase domainFamily: Nitric oxide (NO) synthase oxygenase domain
77 c5lc5m_
not modelled
5.2
13
PDB header: oxidoreductaseChain: M: PDB Molecule: nadh-ubiquinone oxidoreductase chain 4;PDBTitle: structure of mammalian respiratory complex i, class2
78 d1q5uz_
not modelled
5.1
8
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
79 c2rlfA_
not modelled
5.1
25
PDB header: proton transportChain: A: PDB Molecule: matrix protein 2;PDBTitle: proton channel m2 from influenza a in complex with2 inhibitor rimantadine
80 d2r31a1
not modelled
5.1
18
Fold: ATP12-likeSuperfamily: ATP12-likeFamily: ATP12-like