| 1 | c3ukoA_
|
|
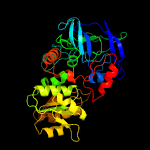 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase class-3;
PDBTitle: crystal structure of s-nitrosoglutathione reductase from arabidopsis2 thaliana, complex with nadh
|
|
|
|
| 2 | c1hf3A_
|
|
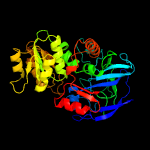 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase e chain;
PDBTitle: atomic x-ray structure of liver alcohol dehydrogenase containing2 cadmium and a hydroxide adduct to nadh
|
|
|
|
| 3 | c5tnxA_
|
|
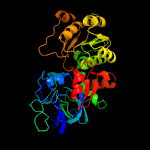 |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase zinc-binding domain protein;
PDBTitle: crystal structure of alcohol dehydrogenase zinc-binding domain protein2 from burkholderia ambifaria
|
|
|
|
| 4 | c4rquA_
|
|
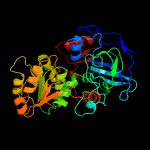 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase crystal structure in complex with nad
|
|
|
|
| 5 | c3krtC_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:crotonyl coa reductase;
PDBTitle: crystal structure of putative crotonyl coa reductase from streptomyces2 coelicolor a3(2)
|
|
|
|
| 6 | c1f8fA_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:benzyl alcohol dehydrogenase;
PDBTitle: crystal structure of benzyl alcohol dehydrogenase from acinetobacter2 calcoaceticus
|
|
|
|
| 7 | c3cosD_
|
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 4;
PDBTitle: crystal structure of human class ii alcohol dehydrogenase (adh4) in2 complex with nad and zn
|
|
|
|
| 8 | c1ma0B_
|
|
 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glutathione-dependent formaldehyde dehydrogenase;
PDBTitle: ternary complex of human glutathione-dependent formaldehyde2 dehydrogenase with nad+ and dodecanoic acid
|
|
|
|
| 9 | c4gi2B_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:crotonyl-coa carboxylase/reductase;
PDBTitle: crotonyl-coa carboxylase/reductase
|
|
|
|
| 10 | c1cdoB_
|
|
 |
100.0 |
33 |
PDB header:oxidoreductase (ch-oh(d)-nad(a))
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase (e.c.1.1.1.1) (ee isozyme) complexed with2 nicotinamide adenine dinucleotide (nad), and zinc
|
|
|
|
| 11 | c1p0fA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: crystal structure of the binary complex: nadp(h)-dependent vertebrate2 alcohol dehydrogenase (adh8) with the cofactor nadp
|
|
|
|
| 12 | c4y0kA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ante;
PDBTitle: structure of crotonyl-coa carboxylase/reductase ante in complex with2 nadp
|
|
|
|
| 13 | c2dphA_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dismutase;
PDBTitle: crystal structure of formaldehyde dismutase
|
|
|
|
| 14 | c4a10A_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:octenoyl-coa reductase/carboxylase;
PDBTitle: apo-structure of 2-octenoyl-coa carboxylase reductase cinf from2 streptomyces sp.
|
|
|
|
| 15 | c1pl6A_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sorbitol dehydrogenase;
PDBTitle: human sdh/nadh/inhibitor complex
|
|
|
|
| 16 | c4a2cB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:galactitol-1-phosphate 5-dehydrogenase;
PDBTitle: crystal structure of galactitol-1-phosphate dehydrogenase from2 escherichia coli
|
|
|
|
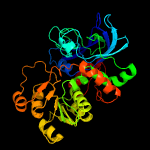
| 17 | c1kolA_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:formaldehyde dehydrogenase;
PDBTitle: crystal structure of formaldehyde dehydrogenase
|
|
|
|
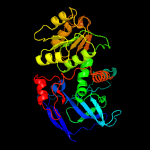
| 18 | c2dfvB_
|
|
 |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable l-threonine 3-dehydrogenase;
PDBTitle: hyperthermophilic threonine dehydrogenase from pyrococcus horikoshii
|
|
|
|
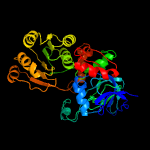
| 19 | c3ip1C_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: structure of putative alcohol dehydrogenase (tm_042) from thermotoga2 maritima
|
|
|
|
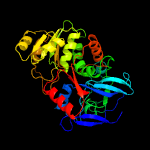
| 20 | c4oaqA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:r-specific carbonyl reductase;
PDBTitle: crystal structure of the r-specific carbonyl reductase from candida2 parapsilosis atcc 7330
|
|
|
|
| 21 | c4ejmA_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative zinc-binding dehydrogenase;
PDBTitle: crystal structure of a putative zinc-binding dehydrogenase (target2 psi-012003) from sinorhizobium meliloti 1021 bound to nadp
|
|
|
| 22 | c1vj0B_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: crystal structure of alcohol dehydrogenase (tm0436) from thermotoga2 maritima at 2.00 a resolution
|
|
|
| 23 | c1r37B_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad-dependent alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase from sulfolobus solfataricus2 complexed with nad(h) and 2-ethoxyethanol
|
|
|
| 24 | c3uogB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehydrogenase from sinorhizobium2 meliloti 1021
|
|
|
| 25 | c1kevB_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: structure of nadp-dependent alcohol dehydrogenase
|
|
|
| 26 | c2hcyD_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 1;
PDBTitle: yeast alcohol dehydrogenase i, saccharomyces cerevisiae fermentative2 enzyme
|
|
|
| 27 | c2cf5A_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cinnamyl alcohol dehydrogenase;
PDBTitle: crystal structures of the arabidopsis cinnamyl alcohol2 dehydrogenases, atcad5
|
|
|
| 28 | c4j6fB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative alcohol dehydrogenase;
PDBTitle: crystal structure of putative alcohol dehydrogenase from sinorhizobium2 meliloti 1021, nysgrc-target 012230
|
|
|
| 29 | c1yqxB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sinapyl alcohol dehydrogenase;
PDBTitle: sinapyl alcohol dehydrogenase at 2.5 angstrom resolution
|
|
|
| 30 | c4cpdA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase tadh from thermus sp. atn1
|
|
|
| 31 | c5k1sD_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase, zinc-binding dehydrogenase family;
PDBTitle: crystal structure of aibc
|
|
|
| 32 | c2eihA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad-dependent alcohol dehydrogenase
|
|
|
| 33 | c4ilkB_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:starvation sensing protein rspb;
PDBTitle: crystal structure of short chain alcohol dehydrogenase (rspb) from e.2 coli cft073 (efi target efi-506413) complexed with cofactor nadh
|
|
|
| 34 | c1h2bA_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the alcohol dehydrogenase from the2 hyperthermophilic archaeon aeropyrum pernix at 1.65a resolution
|
|
|
| 35 | c5kiaA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: crystal structure of l-threonine 3-dehydrogenase from burkholderia2 thailandensis
|
|
|
| 36 | c5vm2A_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of eck1772, an oxidoreductase/dehydrogenase of2 unknown specificity involved in membrane biogenesis from escherichia3 coli
|
|
|
| 37 | c2ouiB_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent alcohol dehydrogenase;
PDBTitle: d275p mutant of alcohol dehydrogenase from protozoa entamoeba2 histolytica
|
|
|
| 38 | c5ylnB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: zinc dependent alcohol dehydrogenase 2 from streptococcus pneumonia -2 apo form
|
|
|
| 39 | c1piwA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:hypothetical zinc-type alcohol dehydrogenase-
PDBTitle: apo and holo structures of an nadp(h)-dependent cinnamyl2 alcohol dehydrogenase from saccharomyces cerevisiae
|
|
|
| 40 | c5vktB_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cinnamyl alcohol dehydrogenases (sbcad4);
PDBTitle: cinnamyl alcohol dehydrogenases (sbcad4) from sorghum bicolor (l.)2 moench
|
|
|
| 41 | c2ejvA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: crystal structure of threonine 3-dehydrogenase complexed with nad+
|
|
|
| 42 | c4eezB_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase 1;
PDBTitle: crystal structure of lactococcus lactis alcohol dehydrogenase variant2 re1
|
|
|
| 43 | c1e3jA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp(h)-dependent ketose reductase;
PDBTitle: ketose reductase (sorbitol dehydrogenase) from silverleaf2 whitefly
|
|
|
| 44 | c1lluD_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase;
PDBTitle: the ternary complex of pseudomonas aeruginosa alcohol2 dehydrogenase with its coenzyme and weak substrate
|
|
|
| 45 | c2xaaC_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:secondary alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase adh-'a' from rhodococcus ruber dsm2 44541 at ph 8.5 in complex with nad and butane-1,4-diol
|
|
|
| 46 | c4gkvC_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:alcohol dehydrogenase, propanol-preferring;
PDBTitle: structure of escherichia coli adhp (ethanol-inducible dehydrogenase)2 with bound nad
|
|
|
| 47 | c6n7lB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of an alcohol dehydrogenase from elizabethkingia2 anophelis nuhp1
|
|
|
| 48 | c6dkhC_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:l-idonate 5-dehydrogenase (nad(p)(+));
PDBTitle: the crystal structure of l-idonate 5-dehydrogenase from escherichia2 coli str. k-12 substr. mg1655
|
|
|
| 49 | c5fi5A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tetrahydroalstonine synthase;
PDBTitle: heteroyohimbine synthase thas1 from catharanthus roseus - apo form
|
|
|
| 50 | c2cdaA_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose dehydrogenase;
PDBTitle: sulfolobus solfataricus glucose dehydrogenase 1 in complex with nadp
|
|
|
| 51 | c3b70A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl reductase;
PDBTitle: crystal structure of aspergillus terreus trans-acting lovastatin2 polyketide enoyl reductase (lovc) with bound nadp
|
|
|
| 52 | c3qwbC_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:probable quinone oxidoreductase;
PDBTitle: crystal structure of saccharomyces cerevisiae zeta-crystallin-like2 quinone oxidoreductase zta1 complexed with nadph
|
|
|
| 53 | c6c49A_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of alcohol dehydrogenase from acinetobacter2 baumannii
|
|
|
| 54 | c4jbiB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alcohol dehydrogenase (zinc);
PDBTitle: 2.35a resolution structure of nadph bound thermostable alcohol2 dehydrogenase from pyrobaculum aerophilum
|
|
|
| 55 | c2j8zA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of human p53 inducible oxidoreductase (2 tp53i3,pig3)
|
|
|
| 56 | c3widC_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:glucose 1-dehydrogenase;
PDBTitle: structure of a glucose dehydrogenase t277f mutant in complex with nadp
|
|
|
| 57 | c5h81A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:heteroyohimbine synthase thas2;
PDBTitle: heteroyohimbine synthase thas2 from catharanthus roseus - complex with2 nadp+
|
|
|
| 58 | c4z6kD_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase;
PDBTitle: alcohol dehydrogenase from the antarctic psychrophile moraxella sp.2 tae 123
|
|
|
| 59 | c1rjwA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of nad(+)-dependent alcohol dehydrogenase2 from bacillus stearothermophilus strain lld-r
|
|
|
| 60 | c1uufA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:zinc-type alcohol dehydrogenase-like protein
PDBTitle: crystal structure of a zinc-type alcohol dehydrogenase-like2 protein yahk
|
|
|
| 61 | c3m6iA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-arabinitol 4-dehydrogenase;
PDBTitle: l-arabinitol 4-dehydrogenase
|
|
|
| 62 | c3pi7A_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh oxidoreductase;
PDBTitle: crystal structure of a putative nadph:quinone reductase (mll3093) from2 mesorhizobium loti at 1.71 a resolution
|
|
|
| 63 | c2vwpA_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose dehydrogenase;
PDBTitle: haloferax mediterranei glucose dehydrogenase in complex with nadph and2 zn.
|
|
|
| 64 | c1n9gF_ |
|
not modelled |
100.0 |
15 |
PDB header:hydrolase
Chain: F: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: mitochondrial 2-enoyl thioester reductase etr1p/etr2p heterodimer from2 candida tropicalis
|
|
|
| 65 | c3gazA_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase superfamily protein;
PDBTitle: crystal structure of an alcohol dehydrogenase superfamily protein from2 novosphingobium aromaticivorans
|
|
|
| 66 | c4rvsA_ |
|
not modelled |
100.0 |
23 |
PDB header:electron transport
Chain: A: PDB Molecule:probable quinone reductase qor (nadph:quinone reductase)
PDBTitle: the native structure of mycobacterial quinone oxidoreductase rv154c.
|
|
|
| 67 | c1qorA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of escherichia coli quinone oxidoreductase complexed2 with nadph
|
|
|
| 68 | c3iupB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative nadph:quinone oxidoreductase;
PDBTitle: crystal structure of putative nadph:quinone oxidoreductase2 (yp_296108.1) from ralstonia eutropha jmp134 at 1.70 a resolution
|
|
|
| 69 | c3fbgA_ |
|
not modelled |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative arginate lyase;
PDBTitle: crystal structure of a putative arginate lyase from staphylococcus2 haemolyticus
|
|
|
| 70 | c2j3iB_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent oxidoreductase p1;
PDBTitle: crystal structure of arabidopsis thaliana double bond2 reductase (at5g16970)-binary complex
|
|
|
| 71 | c4dvjA_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative zinc-dependent alcohol dehydrogenase protein;
PDBTitle: crystal structure of a putative zinc-dependent alcohol dehydrogenase2 protein from rhizobium etli cfn 42
|
|
|
| 72 | c2c0cB_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:zinc binding alcohol dehydrogenase, domain containing 2;
PDBTitle: structure of the mgc45594 gene product
|
|
|
| 73 | c5zxnA_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp-dependent oxidoreductase;
PDBTitle: crystal structure of cura from vibrio vulnificus
|
|
|
| 74 | c1o89A_ |
|
not modelled |
100.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:yhdh;
PDBTitle: crystal structure of e. coli k-12 yhdh
|
|
|
| 75 | c1wlyA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-haloacrylate reductase;
PDBTitle: crystal structure of 2-haloacrylate reductase
|
|
|
| 76 | c1xa0B_ |
|
not modelled |
100.0 |
18 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative nadph dependent oxidoreductases;
PDBTitle: crystal structure of mcsg target apc35536 from bacillus2 stearothermophilus
|
|
|
| 77 | c3gmsA_ |
|
not modelled |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative nadph:quinone reductase;
PDBTitle: crystal structure of putative nadph:quinone reductase from bacillus2 thuringiensis
|
|
|
| 78 | c2vcyA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:trans-2-enoyl-coa reductase;
PDBTitle: crystal structure of 2-enoyl thioester reductase of human fas ii
|
|
|
| 79 | c5gxeA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:acrylyl-coa reductase acui;
PDBTitle: crystal structure of acryloyl-coa reductase acui in complex with nadph
|
|
|
| 80 | c5a3jF_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:putative quinone-oxidoreductase homolog, chloroplastic;
PDBTitle: crystal structure of the chloroplastic gamma-ketol reductase from2 arabidopsis thaliana bound to 13-oxo-9(z),11(e),15(z)-3 octadecatrienoic acid.
|
|
|
| 81 | c3jynA_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structures of pseudomonas syringae pv. tomato dc30002 quinone oxidoreductase complexed with nadph
|
|
|
| 82 | c4jxkA_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase rop_24000 (target efi-506400) from2 rhodococcus opacus b4
|
|
|
| 83 | c1yb5A_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of human zeta-crystallin with bound nadp
|
|
|
| 84 | c2h6eA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-arabinose 1-dehydrogenase;
PDBTitle: crystal structure of the d-arabinose dehydrogenase from sulfolobus2 solfataricus
|
|
|
| 85 | c1y9eB_ |
|
not modelled |
100.0 |
19 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein yhfp;
PDBTitle: crystal structure of bacillus subtilis protein yhfp with nad bound
|
|
|
| 86 | c4b7cB_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase;
PDBTitle: crystal structure of hypothetical protein pa1648 from2 pseudomonas aeruginosa.
|
|
|
| 87 | c3tqhA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: structure of the quinone oxidoreductase from coxiella burnetii
|
|
|
| 88 | c3gohA_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase, zinc-containing;
PDBTitle: crystal structure of alcohol dehydrogenase superfamily protein2 (np_718042.1) from shewanella oneidensis at 1.55 a resolution
|
|
|
| 89 | c6eowC_ |
|
not modelled |
100.0 |
16 |
PDB header:transferase
Chain: C: PDB Molecule:ketone/zingerone synthase 1;
PDBTitle: structure of raspberry ketone synthase with hydroxybenzalacetone
|
|
|
| 90 | c4idbA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ripening-induced protein;
PDBTitle: structure of the fragaria x ananassa enone oxidoreductase in complex2 with nadp+
|
|
|
| 91 | c2w4qA_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prostaglandin reductase 2;
PDBTitle: crystal structure of human zinc-binding alcohol2 dehydrogenase 1 (zadh1) in ternary complex with nadp and3 18beta-glycyrrhetinic acid
|
|
|
| 92 | c4mkrC_ |
|
not modelled |
100.0 |
16 |
PDB header:plant protein
Chain: C: PDB Molecule:zingiber officinale double bond reductase;
PDBTitle: structure of the apo form of a zingiber officinale double bond2 reductase
|
|
|
| 93 | c4dupB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structure of a quinone oxidoreductase from rhizobium etli cfn2 42
|
|
|
| 94 | c4eyeA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable oxidoreductase;
PDBTitle: crystal structure of a probable oxidoreductase from mycobacterium2 abscessus solved by iodide ion sad
|
|
|
| 95 | c5dp2A_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:curf;
PDBTitle: curf er cyclopropanase from curacin a biosynthetic pathway
|
|
|
| 96 | c4a27A_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:synaptic vesicle membrane protein vat-1 homolog-like;
PDBTitle: crystal structure of human synaptic vesicle membrane protein vat-12 homolog-like protein
|
|
|
| 97 | c1zsvB_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadp-dependent leukotriene b4 12-
PDBTitle: crystal structure of human nadp-dependent leukotriene b4 12-2 hydroxydehydrogenase
|
|
|
| 98 | c5dp1A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:curk;
PDBTitle: crystal structure of curk enoyl reductase
|
|
|
| 99 | c3slkB_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:polyketide synthase extender module 2;
PDBTitle: structure of ketoreductase and enoylreductase didomain from modular2 polyketide synthase
|
|
|
| 100 | c5dovB_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:jamj;
PDBTitle: crystal structure of jamj enoyl reductase (apo form)
|
|
|
| 101 | c2vn8B_ |
|
not modelled |
100.0 |
21 |
PDB header:receptor inhibitor
Chain: B: PDB Molecule:reticulon-4-interacting protein 1;
PDBTitle: crystal structure of human reticulon 4 interacting protein 1 in2 complex with nadph
|
|
|
| 102 | c1iz0A_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:quinone oxidoreductase;
PDBTitle: crystal structures of the quinone oxidoreductase from2 thermus thermophilus hb8 and its complex with nadph
|
|
|
| 103 | d1p0fa1 |
|
not modelled |
100.0 |
26 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 104 | d1cdoa1 |
|
not modelled |
100.0 |
28 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 105 | d1pl8a1 |
|
not modelled |
100.0 |
25 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 106 | d1uufa1 |
|
not modelled |
100.0 |
25 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 107 | d1piwa1 |
|
not modelled |
100.0 |
19 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 108 | d1kola1 |
|
not modelled |
100.0 |
21 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 109 | d1d1ta1 |
|
not modelled |
100.0 |
24 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 110 | d1u3ta1 |
|
not modelled |
100.0 |
30 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 111 | d1u3wa1 |
|
not modelled |
100.0 |
28 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 112 | d2jhfa1 |
|
not modelled |
100.0 |
27 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 113 | d1u3ua1 |
|
not modelled |
100.0 |
29 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 114 | d1e3ia1 |
|
not modelled |
100.0 |
27 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 115 | d2fzwa1 |
|
not modelled |
100.0 |
28 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 116 | d1f8fa1 |
|
not modelled |
100.0 |
27 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 117 | d1jqba1 |
|
not modelled |
100.0 |
18 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 118 | d1llua1 |
|
not modelled |
100.0 |
28 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 119 | d1vj0a1 |
|
not modelled |
99.9 |
21 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|
| 120 | d1jvba1 |
|
not modelled |
99.9 |
27 |
Fold:GroES-like
Superfamily:GroES-like
Family:Alcohol dehydrogenase-like, N-terminal domain |
|
|














































































































































































































































