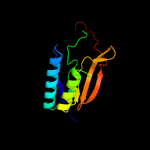
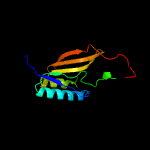
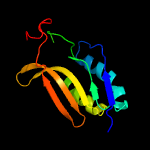
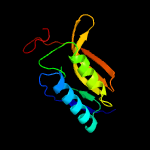
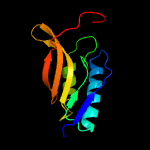
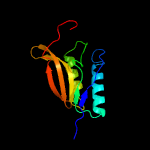
1 c5n6mA_
100.0
24
PDB header: membrane proteinChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
2 c5vrhA_
100.0
24
PDB header: transferaseChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
3 c2w1vA_
99.7
18
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
4 d1uf5a_
99.7
19
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
5 c2e11B_
99.7
22
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
6 c5khaA_
99.7
21
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad+ synthetase;PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
7 d1emsa2
99.7
12
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
8 c5h8lM_
99.7
24
PDB header: hydrolaseChain: M: PDB Molecule: n-carbamoylputrescine amidohydrolase;PDBTitle: crystal structure of medicago truncatula n-carbamoylputrescine2 amidohydrolase (mtcpa) c158s mutant in complex with putrescine
9 c1emsB_
99.7
13
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
10 c3hkxA_
99.7
22
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
11 d1j31a_
99.7
23
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
12 c3wuyA_
99.7
17
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase;PDBTitle: crystal structure of nit6803
13 c5hyyA_
99.7
17
PDB header: hydrolaseChain: A: PDB Molecule: nta1p;PDBTitle: crystal structure of n-terminal amidase
14 c3dlaD_
99.6
24
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
15 c4hg3C_
99.6
18
PDB header: hydrolaseChain: C: PDB Molecule: probable hydrolase nit2;PDBTitle: structural insights into yeast nit2: wild-type yeast nit2 in complex2 with alpha-ketoglutarate
16 c2plqA_
99.6
23
PDB header: hydrolaseChain: A: PDB Molecule: aliphatic amidase;PDBTitle: crystal structure of the amidase from geobacillus pallidus rapc8
17 c2vhiG_
99.6
19
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
18 c6ftqA_
99.6
14
PDB header: hydrolaseChain: A: PDB Molecule: beta-ureidopropionase;PDBTitle: crystal structure of human beta-ureidopropionase (beta-alanine2 synthase) - mutant t299c
19 c4f4hA_
99.6
24
PDB header: ligaseChain: A: PDB Molecule: glutamine dependent nad+ synthetase;PDBTitle: crystal structure of a glutamine dependent nad+ synthetase from2 burkholderia thailandensis
20 c3ilvA_
99.6
12
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
21 d1f89a_
not modelled
99.6
21
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
22 c2vhhA_
not modelled
99.6
19
PDB header: hydrolaseChain: A: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
23 c3n05B_
not modelled
99.6
21
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
24 c6i00C_
not modelled
99.6
25
PDB header: hydrolaseChain: C: PDB Molecule: bifunctional nitrilase/nitrile hydratase nit4;PDBTitle: cryo-em informed directed evolution of nitrilase 4 leads to a change2 in quaternary structure.
25 c2e2kC_
not modelled
99.5
17
PDB header: hydrolaseChain: C: PDB Molecule: formamidase;PDBTitle: helicobacter pylori formamidase amif contains a fine-tuned cysteine-2 glutamate-lysine catalytic triad
26 c6mg6D_
not modelled
99.5
15
PDB header: hydrolaseChain: D: PDB Molecule: carbon-nitrogen hydrolase;PDBTitle: crystal structure of carbon-nitrogen hydrolase from helicobacter2 pylori g27
27 c4cyyA_
not modelled
99.5
20
PDB header: hydrolaseChain: A: PDB Molecule: pantetheinase;PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
28 d1ejia_
not modelled
63.8
26
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
29 c6ic4I_
not modelled
61.5
15
PDB header: protein transportChain: I: PDB Molecule: abc transporter atp-binding protein;PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
30 c3n8hA_
not modelled
59.6
14
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
31 c3gfoA_
not modelled
59.4
11
PDB header: atp binding proteinChain: A: PDB Molecule: cobalt import atp-binding protein cbio 1;PDBTitle: structure of cbio1 from clostridium perfringens: part of the abc2 transporter complex cbionq.
32 d1g2912
not modelled
58.7
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
33 c5y0tD_
not modelled
56.8
11
PDB header: ligaseChain: D: PDB Molecule: thermotoga maritima tmcal;PDBTitle: crystal structure of thermotoga maritima tmcal bound with alpha-thio2 atp(form ii)
34 d1v43a3
not modelled
53.7
12
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
35 c1vciA_
not modelled
53.0
12
PDB header: transport proteinChain: A: PDB Molecule: sugar-binding transport atp-binding protein;PDBTitle: crystal structure of the atp-binding cassette of multisugar2 transporter from pyrococcus horikoshii ot3 complexed with3 atp
36 c3uk2B_
not modelled
50.7
16
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
37 c5b57D_
not modelled
50.6
12
PDB header: metal transportChain: D: PDB Molecule: hemin import atp-binding protein hmuv;PDBTitle: inward-facing conformation of abc heme importer bhuuv from2 burkholderia cenocepacia
38 d1v8fa_
not modelled
47.9
16
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)
39 c4mycC_
not modelled
47.6
8
PDB header: transport proteinChain: C: PDB Molecule: iron-sulfur clusters transporter atm1, mitochondrial;PDBTitle: structure of the mitochondrial abc transporter, atm1
40 c2ejcA_
not modelled
46.5
16
PDB header: ligaseChain: A: PDB Molecule: pantoate--beta-alanine ligase;PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
41 d2awna2
not modelled
46.5
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
42 c5d95A_
not modelled
46.1
17
PDB header: transferaseChain: A: PDB Molecule: aminotransferase class-iii;PDBTitle: structure of thermostable omega-transaminase
43 d1yexa1
not modelled
45.4
16
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione peroxidase-like
44 c1q1bD_
not modelled
45.2
11
PDB header: transport proteinChain: D: PDB Molecule: maltose/maltodextrin transport atp-binding protein malk;PDBTitle: crystal structure of e. coli malk in the nucleotide-free form
45 c4g1uD_
not modelled
45.1
12
PDB header: transport protein/hydrolaseChain: D: PDB Molecule: hemin import atp-binding protein hmuv;PDBTitle: x-ray structure of the bacterial heme transporter hmuuv from yersinia2 pestis
46 c3wmeA_
not modelled
44.6
14
PDB header: transport proteinChain: A: PDB Molecule: atp-binding cassette, sub-family b, member 1;PDBTitle: crystal structure of an inward-facing eukaryotic abc multidrug2 transporter
47 d3dhwc1
not modelled
44.5
14
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
48 c4e77A_
not modelled
44.5
19
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: 2.0a crystal structure of a glutamate-1-semialdehyde aminotransferase2 from yersinia pestis co92
49 c3mxtA_
not modelled
44.1
20
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
50 d1oxxk2
not modelled
43.6
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
51 c3l44A_
not modelled
43.5
14
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase 1;PDBTitle: crystal structure of bacillus anthracis heml-1, glutamate semialdehyde2 aminotransferase
52 c2yyzA_
not modelled
43.3
13
PDB header: transport proteinChain: A: PDB Molecule: sugar abc transporter, atp-binding protein;PDBTitle: crystal structure of sugar abc transporter, atp-binding protein
53 c5y0nB_
not modelled
42.0
13
PDB header: ligaseChain: B: PDB Molecule: upf0348 protein b4417_3650;PDBTitle: crystal structure of bacillus subtilis tmcal bound with atp (semet2 derivative)
54 c3bs8A_
not modelled
40.3
11
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: crystal structure of glutamate 1-semialdehyde aminotransferase2 complexed with pyridoxamine-5'-phosphate from bacillus subtilis
55 c3n0lA_
not modelled
39.5
26
PDB header: transferaseChain: A: PDB Molecule: serine hydroxymethyltransferase;PDBTitle: crystal structure of serine hydroxymethyltransferase from2 campylobacter jejuni
56 c2cy8A_
not modelled
38.7
17
PDB header: transferaseChain: A: PDB Molecule: d-phenylglycine aminotransferase;PDBTitle: crystal structure of d-phenylglycine aminotransferase (d-phgat) from2 pseudomonas strutzeri st-201
57 d1ihoa_
not modelled
37.2
24
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)
58 c2zsmA_
not modelled
35.8
24
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-2 aminomutase from aeropyrum pernix, hexagonal form
59 c4mkiB_
not modelled
35.2
12
PDB header: hydrolaseChain: B: PDB Molecule: energy-coupling factor transporter atp-binding proteinPDBTitle: cobalt transporter atp-binding subunit
60 d1jj7a_
not modelled
35.1
11
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
61 c4fwiB_
not modelled
34.6
13
PDB header: transport proteinChain: B: PDB Molecule: abc-type dipeptide/oligopeptide/nickel transport system,PDBTitle: crystal structure of the nucleotide-binding domain of a dipeptide abc2 transporter
62 c1oxtB_
not modelled
33.3
21
PDB header: transport proteinChain: B: PDB Molecule: abc transporter, atp binding protein;PDBTitle: crystal structure of glcv, the abc-atpase of the glucose abc2 transporter from sulfolobus solfataricus
63 d1l2ta_
not modelled
33.2
13
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
64 c2a7vA_
not modelled
33.1
28
PDB header: transferaseChain: A: PDB Molecule: serine hydroxymethyltransferase;PDBTitle: human mitochondrial serine hydroxymethyltransferase 2
65 d2a7va1
not modelled
33.1
28
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
66 c2it1B_
not modelled
32.8
13
PDB header: transport proteinChain: B: PDB Molecule: 362aa long hypothetical maltose/maltodextrinPDBTitle: structure of ph0203 protein from pyrococcus horikoshii
67 c4hzuB_
not modelled
31.7
13
PDB header: hydrolase, transport proteinChain: B: PDB Molecule: energy-coupling factor transporter atp-binding protein ecfaPDBTitle: structure of a bacterial energy-coupling factor transporter
68 c3i4jC_
not modelled
31.6
8
PDB header: transferaseChain: C: PDB Molecule: aminotransferase, class iii;PDBTitle: crystal structure of aminotransferase, class iii from deinococcus2 radiodurans
69 c1z47B_
not modelled
31.1
8
PDB header: ligand binding proteinChain: B: PDB Molecule: putative abc-transporter atp-binding protein;PDBTitle: structure of the atpase subunit cysa of the putative sulfate atp-2 binding cassette (abc) transporter from alicyclobacillus3 acidocaldarius
70 c3dodA_
not modelled
30.3
12
PDB header: transferaseChain: A: PDB Molecule: adenosylmethionine-8-amino-7-oxononanoate aminotransferase;PDBTitle: crystal structure of plp bound 7,8-diaminopelargonic acid synthase in2 bacillus subtilis
71 d2gsaa_
not modelled
28.8
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
72 c6iz9B_
not modelled
28.2
15
PDB header: transferaseChain: B: PDB Molecule: beta-transaminase;PDBTitle: crystal structure of the apo form of a beta-transaminase from2 mesorhizobium sp. strain luk
73 c5ochH_
not modelled
27.9
17
PDB header: hydrolaseChain: H: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
74 c4uoxB_
not modelled
27.9
11
PDB header: transferaseChain: B: PDB Molecule: putrescine aminotransferase;PDBTitle: crystal structure of ygjg in complex with pyridoxal-5'-phosphate2 and putrescine
75 c2ei9A_
not modelled
27.7
22
PDB header: gene regulationChain: A: PDB Molecule: non-ltr retrotransposon r1bmks orf2 protein;PDBTitle: crystal structure of r1bm endonuclease domain
76 c4hziA_
not modelled
27.6
22
PDB header: transport proteinChain: A: PDB Molecule: abc transporter atp-binding protein;PDBTitle: crystal structure of the leptospira interrogans atpase subunit of an2 orphan abc transporter
77 c5u1dA_
not modelled
27.5
10
PDB header: transport proteinChain: A: PDB Molecule: antigen peptide transporter 1;PDBTitle: cryo-em structure of the human tap atp-binding cassette transporter
78 c3d31B_
not modelled
27.3
10
PDB header: transport proteinChain: B: PDB Molecule: sulfate/molybdate abc transporter, atp-binding protein;PDBTitle: modbc from methanosarcina acetivorans
79 c5viuB_
not modelled
27.1
17
PDB header: transferaseChain: B: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase from2 elizabethkingia anophelis
80 c4yerB_
not modelled
27.0
8
PDB header: hydrolaseChain: B: PDB Molecule: abc transporter atp-binding protein;PDBTitle: crystal structure of an abc transporter atp-binding protein (tm_1403)2 from thermotoga maritima msb8 at 2.35 a resolution
81 c5do7B_
not modelled
26.6
14
PDB header: transport proteinChain: B: PDB Molecule: atp-binding cassette sub-family g member 8;PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
82 c6cd1A_
not modelled
26.5
30
PDB header: transferaseChain: A: PDB Molecule: serine hydroxymethyltransferase;PDBTitle: crystal structure of medicago truncatula serine2 hydroxymethyltransferase 3 (mtshmt3), complexes with reaction3 intermediates
83 c2d62A_
not modelled
25.5
12
PDB header: sugar binding proteinChain: A: PDB Molecule: multiple sugar-binding transport atp-bindingPDBTitle: crystal structure of multiple sugar binding transport atp-2 binding protein
84 c2ordA_
not modelled
24.7
8
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
85 c3lv2A_
not modelled
24.5
24
PDB header: transferaseChain: A: PDB Molecule: adenosylmethionine-8-amino-7-oxononanoate aminotransferase;PDBTitle: crystal structure of mycobacterium tuberculosis 7,8-diaminopelargonic2 acid synthase in complex with substrate analog sinefungin
86 d2pmka1
not modelled
24.3
9
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
87 c2olkD_
not modelled
23.6
13
PDB header: hydrolaseChain: D: PDB Molecule: amino acid abc transporter;PDBTitle: abc protein artp in complex with adp-beta-s
88 c3fvqB_
not modelled
23.6
8
PDB header: hydrolaseChain: B: PDB Molecule: fe(3+) ions import atp-binding protein fbpc;PDBTitle: crystal structure of the nucleotide binding domain fbpc complexed with2 atp
89 c3dhwC_
not modelled
23.6
13
PDB header: membrane protein/hydrolaseChain: C: PDB Molecule: methionine import atp-binding protein metn;PDBTitle: crystal structure of methionine importer metni
90 c2ykyB_
not modelled
23.3
15
PDB header: transferaseChain: B: PDB Molecule: beta-transaminase;PDBTitle: structural determinants of the beta-selectivity of a bacterial2 aminotransferase
91 d2cfba1
not modelled
23.2
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
92 c2nq2C_
not modelled
23.1
18
PDB header: metal transportChain: C: PDB Molecule: hypothetical abc transporter atp-binding proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
93 c4tqvO_
not modelled
22.8
10
PDB header: transport proteinChain: O: PDB Molecule: algs;PDBTitle: crystal structure of a bacterial abc transporter involved in the2 import of the acidic polysaccharide alginate
94 d3d31a2
not modelled
22.4
10
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ABC transporter ATPase domain-like
95 d1bj4a_
not modelled
22.0
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
96 d1rv3a_
not modelled
21.9
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
97 c3jcuO_
not modelled
21.1
21
PDB header: membrane proteinChain: O: PDB Molecule: oxygen-evolving enhancer protein 1, chloroplastic;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
98 c5f15A_
not modelled
21.1
12
PDB header: transferaseChain: A: PDB Molecule: 4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
99 c5g4iA_
not modelled
20.9
11
PDB header: transferaseChain: A: PDB Molecule: phospholyase;PDBTitle: plp-dependent phospholyase a1rdf1 from arthrobacter aurescens tc1