| 1 |
|
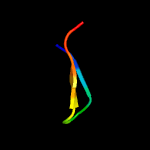
PDB 1eaq chain A
Region: 41 - 57
Aligned: 17
Modelled: 17
Confidence: 41.1%
Identity: 59%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: p53-like transcription factors
Family: RUNT domain
Phyre2
| 2 |
|
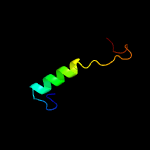
PDB 1ljm chain A
Region: 41 - 57
Aligned: 17
Modelled: 17
Confidence: 38.7%
Identity: 59%
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome f
Superfamily: p53-like transcription factors
Family: RUNT domain
Phyre2
| 3 |
|
PDB 4l0z chain A
Region: 41 - 57
Aligned: 17
Modelled: 17
Confidence: 31.9%
Identity: 59%
PDB header:transcription/dna
Chain: A: PDB Molecule:runt-related transcription factor 1;
PDBTitle: crystal structure of runx1 and ets1 bound to tcr alpha promoter2 (crystal form 2)
Phyre2
| 4 |
|
PDB 2eln chain A
Region: 96 - 109
Aligned: 14
Modelled: 14
Confidence: 15.5%
Identity: 64%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 11th c2h2 zinc finger of human2 zinc finger protein 406
Phyre2
| 5 |
|
PDB 2gjv chain F
Region: 36 - 86
Aligned: 38
Modelled: 51
Confidence: 12.2%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:putative cytoplasmic protein;
PDBTitle: crystal structure of a protein of unknown function from salmonella2 typhimurium
Phyre2
| 6 |
|
PDB 2gjv chain A domain 1
Region: 36 - 86
Aligned: 38
Modelled: 51
Confidence: 12.0%
Identity: 21%
Fold: Phage tail protein-like
Superfamily: Phage tail protein-like
Family: STM4215-like
Phyre2
| 7 |
|
PDB 5ijl chain A
Region: 30 - 50
Aligned: 21
Modelled: 21
Confidence: 10.5%
Identity: 48%
PDB header:transferase
Chain: A: PDB Molecule:dna polymerase ii large subunit;
PDBTitle: d-family dna polymerase - dp2 subunit (catalytic subunit)
Phyre2
| 8 |
|
PDB 2ywx chain A
Region: 25 - 46
Aligned: 18
Modelled: 22
Confidence: 7.9%
Identity: 33%
PDB header:lyase
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase catalytic subunit;
PDBTitle: crystal structure of phosphoribosylaminoimidazole carboxylase2 catalytic subunit from methanocaldococcus jannaschii
Phyre2
| 9 |
|
PDB 4ja0 chain A
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 7.8%
Identity: 33%
PDB header:protein binding
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase;
PDBTitle: crystal structure of the invertebrate bi-functional purine2 biosynthesis enzyme paics at 2.8 a resolution
Phyre2
| 10 |
|
PDB 1xu2 chain R
Region: 97 - 104
Aligned: 8
Modelled: 8
Confidence: 6.8%
Identity: 88%
PDB header:cytokine, hormone/growth factor receptor
Chain: R: PDB Molecule:tumor necrosis factor receptor superfamily member 17;
PDBTitle: the crystal structure of april bound to bcma
Phyre2
| 11 |
|
PDB 1xu2 chain R
Region: 97 - 104
Aligned: 8
Modelled: 8
Confidence: 6.8%
Identity: 88%
Fold: TNF receptor-like
Superfamily: TNF receptor-like
Family: BAFF receptor-like
Phyre2
| 12 |
|
PDB 3ixs chain B
Region: 18 - 31
Aligned: 14
Modelled: 14
Confidence: 6.8%
Identity: 57%
PDB header:protein binding
Chain: B: PDB Molecule:ring1 and yy1-binding protein;
PDBTitle: ring1b c-terminal domain/rybp c-terminal domain complex
Phyre2
| 13 |
|
PDB 3ixs chain F
Region: 18 - 31
Aligned: 14
Modelled: 14
Confidence: 6.7%
Identity: 57%
PDB header:protein binding
Chain: F: PDB Molecule:ring1 and yy1-binding protein;
PDBTitle: ring1b c-terminal domain/rybp c-terminal domain complex
Phyre2
| 14 |
|
PDB 6b3n chain A
Region: 73 - 83
Aligned: 11
Modelled: 11
Confidence: 6.2%
Identity: 73%
PDB header:protein binding
Chain: A: PDB Molecule:nleg5-1;
PDBTitle: solution structure of the n-terminal domain of the effector nleg5-12 from escherichia coli o157:h7 str. sakai
Phyre2
| 15 |
|
PDB 2rps chain A
Region: 95 - 100
Aligned: 6
Modelled: 6
Confidence: 5.9%
Identity: 83%
PDB header:immune system
Chain: A: PDB Molecule:chemokine;
PDBTitle: solution structure of a novel insect chemokine isolated from2 integument
Phyre2
| 16 |
|
PDB 2kn1 chain A
Region: 97 - 104
Aligned: 8
Modelled: 8
Confidence: 5.9%
Identity: 88%
PDB header:signaling protein
Chain: A: PDB Molecule:tumor necrosis factor receptor superfamily member 17;
PDBTitle: solution nmr structure of bcma
Phyre2
| 17 |
|
PDB 2dl0 chain A
Region: 76 - 110
Aligned: 35
Modelled: 35
Confidence: 5.4%
Identity: 31%
PDB header:signaling protein
Chain: A: PDB Molecule:sam and sh3 domain-containing protein 1;
PDBTitle: solution structure of the sam-domain of the sam and sh32 domain containing protein 1
Phyre2
| 18 |
|
PDB 3rgg chain D
Region: 25 - 39
Aligned: 15
Modelled: 15
Confidence: 5.4%
Identity: 20%
PDB header:lyase
Chain: D: PDB Molecule:phosphoribosylaminoimidazole carboxylase, pure protein;
PDBTitle: crystal structure of treponema denticola pure bound to air
Phyre2
| 19 |
|
PDB 3bk3 chain C
Region: 98 - 109
Aligned: 12
Modelled: 12
Confidence: 5.1%
Identity: 42%
PDB header:hormone/growth factor
Chain: C: PDB Molecule:crossveinless 2;
PDBTitle: crystal structure of the complex of bmp-2 and the first von2 willebrand domain type c of crossveinless-2
Phyre2