1 c3jbrE_
55.2
7
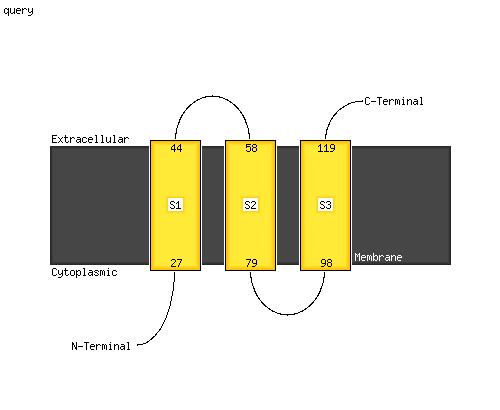
PDB header: membrane proteinChain: E: PDB Molecule: voltage-dependent calcium channel gamma-1 subunit;PDBTitle: cryo-em structure of the rabbit voltage-gated calcium channel cav1.12 complex at 4.2 angstrom
2 c4hr1A_
39.9
45
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of pav1-137, a protein from the virus pav1 that infects2 pyrococcus abyssi.
3 c4ymkA_
34.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa desaturase 1;PDBTitle: crystal structure of stearoyl-coenzyme a desaturase 1
4 c2yevB_
29.6
3
PDB header: electron transportChain: B: PDB Molecule: cytochrome c oxidase subunit 2;PDBTitle: structure of caa3-type cytochrome oxidase
5 c6c4vA_
29.5
58
PDB header: transport proteinChain: A: PDB Molecule: polyketide synthase pks13;PDBTitle: 1.9 angstrom resolution crystal structure of acyl carrier protein2 domain (residues 1350-1461) of polyketide synthase pks13 from3 mycobacterium tuberculosis
6 c5ldxZ_
18.6
21
PDB header: oxidoreductaseChain: Z: PDB Molecule: nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunitPDBTitle: structure of mammalian respiratory complex i, class3.
7 c5lc5Z_
18.6
21
PDB header: oxidoreductaseChain: Z: PDB Molecule: nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunitPDBTitle: structure of mammalian respiratory complex i, class2
8 c5ldwZ_
18.6
21
PDB header: oxidoreductaseChain: Z: PDB Molecule: nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunitPDBTitle: structure of mammalian respiratory complex i, class1
9 d1rh5b_
18.3
8
Fold: Single transmembrane helixSuperfamily: Preprotein translocase SecE subunitFamily: Preprotein translocase SecE subunit
10 d1o4ua2
16.0
35
Fold: alpha/beta-HammerheadSuperfamily: Nicotinate/Quinolinate PRTase N-terminal domain-likeFamily: NadC N-terminal domain-like
11 c6qj4E_
15.8
80
PDB header: cell cycleChain: E: PDB Molecule: condensin complex subunit 2;PDBTitle: crystal structure of the c. thermophilum condensin ycs4-brn12 subcomplex bound to the smc4 atpase head in complex with the c-3 terminal domain of brn1
12 c2l0gA_
14.1
36
PDB header: protein bindingChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution nmr structure of ubiquitin-binding motif (ubm2) of human2 polymerase iota
13 c2khuA_
12.0
36
PDB header: transferase/protein bindingChain: A: PDB Molecule: immunoglobulin g-binding protein g, dnaPDBTitle: solution structure of the ubiquitin-binding motif of human2 polymerase iota
14 d1p4ea1
11.9
50
Fold: SAM domain-likeSuperfamily: lambda integrase-like, N-terminal domainFamily: lambda integrase-like, N-terminal domain
15 c3mk7F_
11.0
12
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
16 c6an7D_
10.3
4
PDB header: transport proteinChain: D: PDB Molecule: transport permease protein;PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
17 c2mc7A_
9.7
10
PDB header: membrane proteinChain: A: PDB Molecule: regulatory peptide;PDBTitle: structure of salmonella mgtr
18 d2ipqx1
9.5
40
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: STY4665 C-terminal domain-like
19 d1v97a2
9.4
33
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
20 c3g43F_
9.2
50
PDB header: metal binding proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel subunit alpha-1c;PDBTitle: crystal structure of the calmodulin-bound cav1.2 c-terminal regulatory2 domain dimer
21 c6gcsW_
not modelled
8.9
18
PDB header: oxidoreductaseChain: W: PDB Molecule: nb6m subunit;PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
22 c2mkvA_
not modelled
8.8
11
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
23 c2vziB_
not modelled
8.4
28
PDB header: cell adhesionChain: B: PDB Molecule: alpha-parvin;PDBTitle: crystal structure of the c-terminal calponin homology domain of alpha-2 parvin in complex with paxillin ld4 motif
24 c2kwuA_
not modelled
8.2
23
PDB header: protein binding/signaling proteinChain: A: PDB Molecule: dna polymerase iota;PDBTitle: solution structure of ubm2 of murine polymerase iota in complex with2 ubiquitin
25 d1s7ba_
not modelled
8.0
14
Fold: Multidrug resistance efflux transporter EmrESuperfamily: Multidrug resistance efflux transporter EmrEFamily: Multidrug resistance efflux transporter EmrE
26 c1fftG_
not modelled
7.8
12
PDB header: oxidoreductaseChain: G: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
27 c5o31Z_
not modelled
7.5
21
PDB header: oxidoreductaseChain: Z: PDB Molecule: nadh dehydrogenase [ubiquinone] 1 alpha subcomplex subunitPDBTitle: mitochondrial complex i in the deactive state
28 c3oxqF_
not modelled
7.5
50
PDB header: metal binding protein/transport proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channel subunit alpha-1c;PDBTitle: crystal structure of ca2+/cam-cav1.2 pre-iq/iq domain complex
29 c2jo1A_
not modelled
7.2
25
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
30 c6cfwI_
not modelled
7.1
8
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
31 c5xyiY_
not modelled
6.8
43
PDB header: ribosomeChain: Y: PDB Molecule: ribosomal protein s24e, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
32 c3shgB_
not modelled
6.5
33
PDB header: transferase/protein bindingChain: B: PDB Molecule: vbha;PDBTitle: vbht fic protein from bartonella schoenbuchensis in complex with vbha2 antitoxin
33 c1al0B_
not modelled
6.5
39
PDB header: virusChain: B: PDB Molecule: scaffolding protein gpb;PDBTitle: procapsid of bacteriophage phix174
34 c2retE_
not modelled
6.4
25
PDB header: protein transportChain: E: PDB Molecule: pseudopilin epsi;PDBTitle: the crystal structure of a binary complex of two pseudopilins: epsi2 and epsj from the type 2 secretion system of vibrio vulnificus
35 c5lnkq_
not modelled
6.4
21
PDB header: oxidoreductaseChain: Q: PDB Molecule: PDBTitle: entire ovine respiratory complex i
36 d1pdaa2
not modelled
6.3
13
Fold: dsRBD-likeSuperfamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domainFamily: Porphobilinogen deaminase (hydroxymethylbilane synthase), C-terminal domain
37 d1jroa2
not modelled
6.1
25
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin domains from multidomain proteins
38 d1dxxa1
not modelled
6.1
23
Fold: CH domain-likeSuperfamily: Calponin-homology domain, CH-domainFamily: Calponin-homology domain, CH-domain
39 d2reta1
not modelled
5.9
25
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: GSPII I/J protein-like
40 d1qlec_
not modelled
5.8
13
Fold: Cytochrome c oxidase subunit III-likeSuperfamily: Cytochrome c oxidase subunit III-likeFamily: Cytochrome c oxidase subunit III-like
41 c6nd1A_
not modelled
5.7
14
PDB header: protein transportChain: A: PDB Molecule: protein translocation protein sec63;PDBTitle: cryoem structure of the sec complex from yeast
42 c3wwtB_
not modelled
5.6
44
PDB header: transcription/viral proteinChain: B: PDB Molecule: protein c';PDBTitle: crystal structure of the y3:stat1nd complex
43 c2k48A_
not modelled
5.2
40
PDB header: viral proteinChain: A: PDB Molecule: nucleoprotein;PDBTitle: nmr structure of the n-terminal coiled coil domain of the2 andes hantavirus nucleocapsid protein
44 c3m7kA_
not modelled
5.2
100
PDB header: hydrolase/dnaChain: A: PDB Molecule: restriction endonuclease paci;PDBTitle: crystal structure of paci-dna enzyme product complex
45 d1qcrd2
not modelled
5.2
80
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain