1 c2f8xM_
53.2
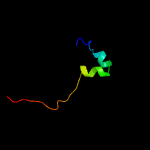
82
PDB header: transcription/dnaChain: M: PDB Molecule: mastermind-like protein 1;PDBTitle: crystal structure of activated notch, csl and maml on hes-12 promoter dna sequence
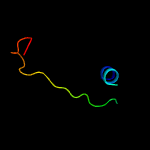
2 d2p10a1
42.0
39
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Mll9387-like
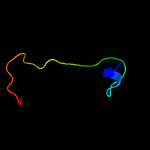
3 c2p10D_
41.9
39
PDB header: hydrolaseChain: D: PDB Molecule: mll9387 protein;PDBTitle: crystal structure of a putative phosphonopyruvate hydrolase (mll9387)2 from mesorhizobium loti maff303099 at 2.15 a resolution
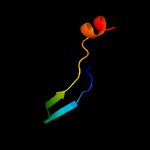
4 d1wgea1
33.2
38
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger
5 c2nudD_
31.5
100
PDB header: toxin/protein bindingChain: D: PDB Molecule: rpm1-interacting protein 4;PDBTitle: the structure of the type iii effector avrb complexed with2 a high-affinity rin4 peptide
6 c2nudC_
31.5
100
PDB header: toxin/protein bindingChain: C: PDB Molecule: rpm1-interacting protein 4;PDBTitle: the structure of the type iii effector avrb complexed with2 a high-affinity rin4 peptide
7 c2jr7A_
29.0
43
PDB header: metal binding proteinChain: A: PDB Molecule: dph3 homolog;PDBTitle: solution structure of human desr1
8 d1ywsa1
28.5
38
Fold: Rubredoxin-likeSuperfamily: CSL zinc fingerFamily: CSL zinc finger
9 c1xnlA_
24.9
44
PDB header: viral proteinChain: A: PDB Molecule: membrane protein gp37;PDBTitle: aslv fusion peptide
10 c5wb8B_
24.5
78
PDB header: signaling proteinChain: B: PDB Molecule: epigen;PDBTitle: crystal structure of the epidermal growth factor receptor2 extracellular region in complex with epigen
11 d1h4ua2
23.0
67
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-like domain of nidogen-1
12 d3pmga4
22.8
37
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain
13 d1kfia4
21.9
37
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain
14 c4i3rG_
19.2
46
PDB header: viral protein/immune systemChain: G: PDB Molecule: outer domain of hiv-1 gp120 (ker2018 od4.2.2);PDBTitle: crystal structure of the outer domain of hiv-1 gp120 in complex with2 vrc-pg04 space group p3221
15 c3j3iA_
18.9
56
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: penicillium chrysogenum virus (pcv) capsid structure
16 d5ruba1
15.7
21
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
17 c9rubB_
14.8
25
PDB header: lyase(carbon-carbon)Chain: B: PDB Molecule: ribulose-1,5-bisphosphate carboxylase;PDBTitle: crystal structure of activated ribulose-1,5-bisphosphate carboxylase2 complexed with its substrate, ribulose-1,5-bisphosphate
18 c3ctrA_
14.6
23
PDB header: hydrolaseChain: A: PDB Molecule: poly(a)-specific ribonuclease parn;PDBTitle: crystal structure of the rrm-domain of the poly(a)-specific2 ribonuclease parn bound to m7gtp
19 c3dnoC_
13.5
50
PDB header: viral proteinChain: C: PDB Molecule: hiv-1 envelope glycoprotein gp120;PDBTitle: molecular structure for the hiv-1 gp120 trimer in the cd4-bound state
20 d1ixsa_
13.1
50
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain
21 d2gp4a1
not modelled
12.9
25
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like
22 c6m8rL_
not modelled
12.6
35
PDB header: protein bindingChain: L: PDB Molecule: gamma-aminobutyric acid type b receptor subunit 2;PDBTitle: crystal structure of the kctd16 btb domain in complex with gabab22 peptide
23 c6m8rK_
not modelled
12.4
35
PDB header: protein bindingChain: K: PDB Molecule: gamma-aminobutyric acid type b receptor subunit 2;PDBTitle: crystal structure of the kctd16 btb domain in complex with gabab22 peptide
24 c2crcA_
not modelled
12.3
50
PDB header: ligaseChain: A: PDB Molecule: ubiquitin conjugating enzyme 7 interactingPDBTitle: solution structure of the zf-ranbp domain of the protein2 hbv associated factor
25 c1h4uA_
not modelled
12.0
43
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
26 c5v8wG_
not modelled
11.7
57
PDB header: hydrolaseChain: G: PDB Molecule: integrator complex subunit 9;PDBTitle: crystal structure of human integrator ints9-ints11 ctd complex
27 c2rusB_
not modelled
11.3
21
PDB header: lyase(carbon-carbon)Chain: B: PDB Molecule: rubisco (ribulose-1,5-bisphosphate carboxylase(slash)PDBTitle: crystal structure of the ternary complex of ribulose-1,5-bisphosphate2 carboxylase, mg(ii), and activator co2 at 2.3-angstroms resolution
28 c5dn8A_
not modelled
10.6
13
PDB header: gtp-binding proteinChain: A: PDB Molecule: gtpase der;PDBTitle: 1.76 angstrom crystal structure of gtp-binding protein der from2 coxiella burnetii in complex with gdp.
29 c1kioA_
not modelled
10.5
47
PDB header: hydrolaseChain: A: PDB Molecule: serine protease inhibitor i;PDBTitle: solution structure of the small serine protease inhibitor2 sgci[l30r, k31m]
30 c2l6lA_
not modelled
10.5
75
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily c member 24;PDBTitle: solution structure of human j-protein co-chaperone, dph4
31 d1fo8a_
not modelled
10.5
71
Fold: Nucleotide-diphospho-sugar transferasesSuperfamily: Nucleotide-diphospho-sugar transferasesFamily: N-acetylglucosaminyltransferase I
32 c5e24D_
not modelled
10.1
22
PDB header: transport/dna binding/dnaChain: D: PDB Molecule: protein hairless;PDBTitle: structure of the su(h)-hairless-dna repressor complex
33 c2mpjA_
not modelled
10.0
55
PDB header: nucleotide binding proteinChain: A: PDB Molecule: recql4-helicase-like protein;PDBTitle: nmr structure of xenopus recq4 zinc knuckle
34 c4djzH_
not modelled
9.8
39
PDB header: hydrolase/hydrolase inhibitorChain: H: PDB Molecule: protease inhibitor sgpi-2;PDBTitle: catalytic fragment of masp-1 in complex with its specific inhibitor2 developed by directed evolution on sgci scaffold
35 c5a48B_
not modelled
9.4
30
PDB header: protein bindingChain: B: PDB Molecule: maternal effect protein oskar;PDBTitle: crystal structure of the lotus domain (aa 139-240) of drosophila2 oskar in p65
36 d1mjsa_
not modelled
9.3
53
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain
37 c5c2gD_
not modelled
9.3
31
PDB header: lyaseChain: D: PDB Molecule: form ii rubisco;PDBTitle: gws1b rubisco: form ii rubisco derived from uncultivated2 gallionellacea species (cabp-bound).
38 c2zkqn_
not modelled
9.2
50
PDB header: ribosomal protein/rnaChain: N: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
39 c3ngbI_
not modelled
9.1
50
PDB header: viral protein/immune systemChain: I: PDB Molecule: envelope glycoprotein gp160;PDBTitle: crystal structure of broadly and potently neutralizing antibody vrc012 in complex with hiv-1 gp120
40 c5te4G_
not modelled
8.9
50
PDB header: immune systemChain: G: PDB Molecule: hiv-1 clade g strain x2088 gp120;PDBTitle: crystal structure of broadly neutralizing vrc01-class antibody n6 in2 complex with hiv-1 clade g strain x2088 gp120 core
41 c2yt2A_
not modelled
8.6
43
PDB header: signaling proteinChain: A: PDB Molecule: fibroblast growth factor receptor substrate 3 and alkPDBTitle: solution structure of the chimera of the ptb domain of snt-2 and 19-2 residue peptide (aa 1571-1589) of halk
42 c5zwoW_
not modelled
8.5
14
PDB header: splicingChain: W: PDB Molecule: 23 kda u4/u6.u5 small nuclear ribonucleoprotein component;PDBTitle: cryo-em structure of the yeast b complex at average resolution of 3.92 angstrom
43 d1lm6a_
not modelled
8.4
75
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase
44 c3tbmA_
not modelled
8.3
50
PDB header: metal binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a type 4 cdgsh iron-sulfur protein.
45 c4r9pA_
not modelled
8.3
40
PDB header: transcriptionChain: A: PDB Molecule: re28239p;PDBTitle: an expansion to the smad mh2-family: the structure of the n-mh22 expanded domain
46 c5a49I_
not modelled
8.2
24
PDB header: protein bindingChain: I: PDB Molecule: maternal effect protein oskar;PDBTitle: crystal structure of the lotus domain (aa 139-222) of2 drosophila oskar in c222
47 c3b08H_
not modelled
8.2
71
PDB header: signaling protein/metal binding proteinChain: H: PDB Molecule: ranbp-type and c3hc4-type zinc finger-containing protein 1;PDBTitle: crystal structure of the mouse hoil1-l-nzf in complex with linear di-2 ubiquitin
48 c1kqiA_
not modelled
8.1
50
PDB header: toxinChain: A: PDB Molecule: actx-hi:ob4219;PDBTitle: nmr solution structure of the trans pro30 isomer of actx-2 hi:ob4219
49 c6ohyA_
not modelled
8.1
50
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein gp160;PDBTitle: chimpanzee siv env trimeric ectodomain.
50 c3l87A_
not modelled
8.0
78
PDB header: hydrolaseChain: A: PDB Molecule: peptide deformylase;PDBTitle: the crystal structure of smu.143c from streptococcus mutans ua159
51 c2hjgA_
not modelled
8.0
20
PDB header: hydrolaseChain: A: PDB Molecule: gtp-binding protein enga;PDBTitle: the crystal structure of the b. subtilis yphc gtpase in complex with2 gdp
52 d1kqha_
not modelled
7.8
50
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins
53 c3j38P_
not modelled
7.6
36
PDB header: ribosomeChain: P: PDB Molecule: 40s ribosomal protein s15, isoform a;PDBTitle: structure of the d. melanogaster 40s ribosomal proteins
54 d1ykwa1
not modelled
7.6
34
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
55 c4kp2A_
not modelled
7.5
41
PDB header: lyaseChain: A: PDB Molecule: homoaconitase large subunit;PDBTitle: crystal structure of homoaconitase large subunit from methanococcus2 jannaschii (mj1003)
56 d2i9aa1
not modelled
7.4
47
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-type module
57 c5vbnB_
not modelled
7.2
27
PDB header: transferaseChain: B: PDB Molecule: dna polymerase epsilon catalytic subunit a;PDBTitle: crystal structure of human dna polymerase epsilon b-subunit in complex2 with c-terminal domain of catalytic subunit
58 d1yymg1
not modelled
7.2
50
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core
59 c4nasD_
not modelled
7.2
28
PDB header: lyaseChain: D: PDB Molecule: ribulose-bisphosphate carboxylase;PDBTitle: the crystal structure of a rubisco-like protein (mtnw) from2 alicyclobacillus acidocaldarius subsp. acidocaldarius dsm 446
60 c3j8gX_
not modelled
7.2
30
PDB header: ribosomeChain: X: PDB Molecule: gtpase der;PDBTitle: electron cryo-microscopy structure of enga bound with the 50s2 ribosomal subunit
61 d1klil_
not modelled
7.1
46
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: EGF/LamininFamily: EGF-type module
62 d1a8da2
not modelled
7.1
30
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Clostridium neurotoxins, C-terminal domain
63 c2bf1A_
not modelled
7.0
42
PDB header: virus proteinChain: A: PDB Molecule: exterior membrane glycoprotein gp120;PDBTitle: structure of an unliganded and fully-glycosylated siv gp120 envelope2 glycoprotein
64 c3bvoA_
not modelled
6.9
29
PDB header: chaperoneChain: A: PDB Molecule: co-chaperone protein hscb, mitochondrial precursor;PDBTitle: crystal structure of human co-chaperone protein hscb
65 d1dd1a_
not modelled
6.8
39
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain
66 c6iicD_
not modelled
6.7
63
PDB header: virusChain: D: PDB Molecule: vp4 of mud crab dicistrovirus;PDBTitle: cryoem structure of mud crab dicistrovirus
67 c4nclB_
not modelled
6.7
18
PDB header: translationChain: B: PDB Molecule: eukaryotic translation initiation factor 5b-like protein;PDBTitle: crystal structure of eukaryotic translation initiation factor eif5b2 (517-970) from chaetomium thermophilum in complex with gdp
68 c3tvjI_
not modelled
6.6
33
PDB header: hydrolaseChain: I: PDB Molecule: protease inhibitor sgpi-2;PDBTitle: catalytic fragment of masp-2 in complex with its specific inhibitor2 developed by directed evolution on sgci scaffold
69 c3ocaB_
not modelled
6.6
63
PDB header: hydrolaseChain: B: PDB Molecule: peptide deformylase;PDBTitle: crystal structure of peptide deformylase from ehrlichia chaffeensis
70 d1lm4a_
not modelled
6.6
78
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase
71 d1ygsa_
not modelled
6.5
39
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain
72 d1jhna4
not modelled
6.4
44
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Calnexin/calreticulin
73 c4kp1A_
not modelled
6.4
46
PDB header: isomeraseChain: A: PDB Molecule: isopropylmalate/citramalate isomerase large subunit;PDBTitle: crystal structure of ipm isomerase large subunit from methanococcus2 jannaschii (mj0499)
74 c3g6nA_
not modelled
6.3
78
PDB header: hydrolaseChain: A: PDB Molecule: peptide deformylase;PDBTitle: crystal structure of an efpdf complex with met-ala-ser
75 c4uzjB_
not modelled
6.2
25
PDB header: hydrolaseChain: B: PDB Molecule: notum;PDBTitle: structure of the wnt deacylase notum from drosophila -2 crystal form i - 2.4a
76 d1g7sa4
not modelled
6.2
19
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: G proteins
77 d1geha1
not modelled
6.2
24
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
78 c5j84A_
not modelled
6.1
36
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase;PDBTitle: crystal structure of l-arabinonate dehydratase in holo-form
79 d1rbla1
not modelled
6.1
34
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
80 c3jwdA_
not modelled
6.1
50
PDB header: viral proteinChain: A: PDB Molecule: hiv-1 gp120 envelope glycoprotein;PDBTitle: structure of hiv-1 gp120 with gp41-interactive region: layered2 architecture and basis of conformational mobility
81 d1khxa_
not modelled
6.1
44
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain
82 c1khxA_
not modelled
6.1
44
PDB header: transcriptionChain: A: PDB Molecule: smad2;PDBTitle: crystal structure of a phosphorylated smad2
83 d1lqya_
not modelled
6.1
78
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase
84 d1wfla_
not modelled
6.0
50
Fold: AN1-like Zinc fingerSuperfamily: AN1-like Zinc fingerFamily: AN1-like Zinc finger
85 d1khua_
not modelled
6.0
47
Fold: SMAD/FHA domainSuperfamily: SMAD/FHA domainFamily: SMAD domain
86 d1ju8a_
not modelled
5.9
60
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Albumin 1
87 c6gzuA_
not modelled
5.9
42
PDB header: transferaseChain: A: PDB Molecule: conserved membrane protein;PDBTitle: structure of chlamydia abortus effector protein chladub
88 c3g5pB_
not modelled
5.8
44
PDB header: hydrolaseChain: B: PDB Molecule: peptide deformylase, mitochondrial;PDBTitle: structure and activity of human mitochondrial peptide deformylase, a2 novel cancer target
89 c3rg0A_
not modelled
5.7
67
PDB header: chaperoneChain: A: PDB Molecule: calreticulin;PDBTitle: structural and functional relationships between the lectin and arm2 domains of calreticulin
90 d1xeoa1
not modelled
5.7
63
Fold: Peptide deformylaseSuperfamily: Peptide deformylaseFamily: Peptide deformylase
91 c3r0rA_
not modelled
5.7
36
PDB header: virusChain: A: PDB Molecule: porcine circovirus 2 (pcv2) capsid protein;PDBTitle: the 2.3 a structure of porcine circovirus 2
92 c1zy1B_
not modelled
5.7
100
PDB header: hydrolaseChain: B: PDB Molecule: peptide deformylase, mitochondrial;PDBTitle: x-ray structure of peptide deformylase from arabidopsis2 thaliana (atpdf1a) in complex with met-ala-ser
93 c5hcfF_
not modelled
5.6
56
PDB header: chaperoneChain: F: PDB Molecule: calreticulin, putative,calreticulin, putative;PDBTitle: t. cruzi calreticulin globular domain
94 c3fk4A_
not modelled
5.5
14
PDB header: isomeraseChain: A: PDB Molecule: rubisco-like protein;PDBTitle: crystal structure of rubisco-like protein from bacillus2 cereus atcc 14579
95 d1bwva1
not modelled
5.5
34
Fold: TIM beta/alpha-barrelSuperfamily: RuBisCo, C-terminal domainFamily: RuBisCo, large subunit, C-terminal domain
96 c5tpjA_
not modelled
5.5
29
PDB header: de novo proteinChain: A: PDB Molecule: denovo ntf2;PDBTitle: crystal structure of a de novo designed protein with curved beta-sheet
97 c3uwaA_
not modelled
5.4
63
PDB header: hydrolaseChain: A: PDB Molecule: riia-riib membrane-associated protein;PDBTitle: crystal structure of a probable peptide deformylase from synechococcus2 phage s-ssm7
98 c5hcaB_
not modelled
5.4
56
PDB header: sugar binding proteinChain: B: PDB Molecule: calreticulin,calreticulin;PDBTitle: globular domain of the entamoeba histolytica calreticulin in complex2 with glucose
99 d2b4cg1
not modelled
5.4
50
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core