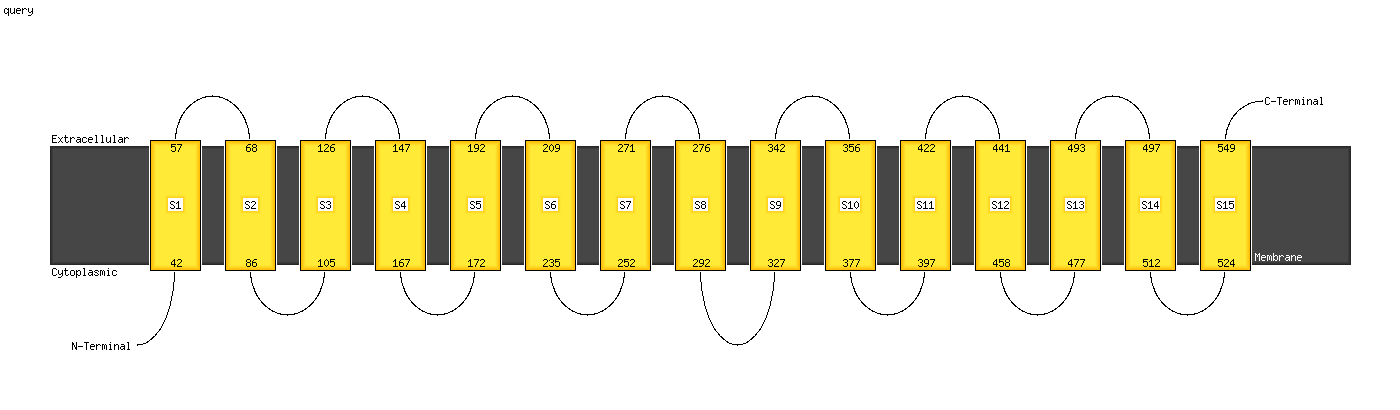
1 c2w2eA_
73.1
13
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin pip2-7 7;PDBTitle: 1.15 angstrom crystal structure of p.pastoris aquaporin, aqy1, in a2 closed conformation at ph 3.5
2 d1j4na_
64.2
17
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like
3 c4r0cB_
61.9
11
PDB header: membrane proteinChain: B: PDB Molecule: abgt putative transporter family;PDBTitle: crystal structure of the alcanivorax borkumensis ydah transporter2 reveals an unusual topology
4 d1fx8a_
40.6
17
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like
5 c6eu6A_
34.0
15
PDB header: membrane proteinChain: A: PDB Molecule: ammonium transporter;PDBTitle: sensor amt protein
6 c2ia2D_
25.1
15
PDB header: transcriptionChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: the crystal structure of a putative transcriptional regulator rha061952 from rhodococcus sp. rha1
7 c1ldaA_
23.4
17
PDB header: transport proteinChain: A: PDB Molecule: glycerol uptake facilitator protein;PDBTitle: crystal structure of the e. coli glycerol facilitator (glpf) without2 substrate glycerol
8 c2b6pA_
20.0
14
PDB header: membrane proteinChain: A: PDB Molecule: lens fiber major intrinsic protein;PDBTitle: x-ray structure of lens aquaporin-0 (aqp0) (lens mip) in an open pore2 state
9 c4g1uB_
17.9
18
PDB header: transport protein/hydrolaseChain: B: PDB Molecule: hemin transport system permease protein hmuu;PDBTitle: x-ray structure of the bacterial heme transporter hmuuv from yersinia2 pestis
10 c2g7uB_
17.7
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: transcriptional regulator;PDBTitle: 2.3 a structure of putative catechol degradative operon regulator from2 rhodococcus sp. rha1
11 c2bruC_
17.4
33
PDB header: oxidoreductaseChain: C: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: complex of the domain i and domain iii of escherichia coli2 transhydrogenase
12 c2nuuF_
17.3
19
PDB header: transport protein/signaling proteinChain: F: PDB Molecule: ammonia channel;PDBTitle: regulating the escherichia coli ammonia channel: the crystal structure2 of the amtb-glnk complex
13 d1d4oa_
15.9
42
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
14 c1pt9B_
15.8
42
PDB header: oxidoreductaseChain: B: PDB Molecule: nad(p) transhydrogenase, mitochondrial;PDBTitle: crystal structure analysis of the diii component of transhydrogenase2 with a thio-nicotinamide nucleotide analogue
15 d1pnoa_
15.6
42
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Transhydrogenase domain III (dIII)
16 c3r4kD_
15.2
15
PDB header: dna binding proteinChain: D: PDB Molecule: transcriptional regulator, iclr family;PDBTitle: crystal structure of a putative iclr transcriptional regulator2 (tm1040_3717) from silicibacter sp. tm1040 at 2.46 a resolution
17 c1mkmA_
13.8
22
PDB header: transcriptionChain: A: PDB Molecule: iclr transcriptional regulator;PDBTitle: crystal structure of the thermotoga maritima iclr
18 c2f2bA_
13.3
16
PDB header: membrane proteinChain: A: PDB Molecule: aquaporin aqpm;PDBTitle: crystal structure of integral membrane protein aquaporin aqpm at 1.68a2 resolution
19 c2xroE_
13.2
30
PDB header: dna-binding protein/dnaChain: E: PDB Molecule: hth-type transcriptional regulator ttgv;PDBTitle: crystal structure of ttgv in complex with its dna operator
20 d2qapa1
12.4
31
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
21 c2qapC_
not modelled
12.2
31
PDB header: lyaseChain: C: PDB Molecule: fructose-1,6-bisphosphate aldolase;PDBTitle: fructose-1,6-bisphosphate aldolase from leishmania mexicana
22 c6c6lN_
not modelled
12.1
16
PDB header: membrane proteinChain: N: PDB Molecule: v0 assembly protein 1;PDBTitle: yeast vacuolar atpase vo in lipid nanodisc
23 c5tjjA_
not modelled
12.0
15
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, iclr family;PDBTitle: crystal structure of icir transcriptional regulator from2 alicyclobacillus acidocaldarius
24 c3c02A_
not modelled
11.9
15
PDB header: membrane proteinChain: A: PDB Molecule: aquaglyceroporin;PDBTitle: x-ray structure of the aquaglyceroporin from plasmodium falciparum
25 c2kncA_
not modelled
11.4
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
26 c5h1aC_
not modelled
10.8
30
PDB header: transcription regulatorChain: C: PDB Molecule: iclr transcription factor homolog;PDBTitle: crystal structure of an iclr homolog from microbacterium sp. strain2 hm58-2
27 c2o0yB_
not modelled
10.3
11
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of putative transcriptional regulator rha1_ro069532 (iclr-family) from rhodococcus sp.
28 c5whmB_
not modelled
10.2
22
PDB header: transcriptionChain: B: PDB Molecule: iclr family transcriptional regulator;PDBTitle: crystal structure of iclr family transcriptional regulator from2 brucella abortus
29 c3mmtC_
not modelled
10.1
33
PDB header: hydrolaseChain: C: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose bisphosphate aldolase from bartonella2 henselae, bound to fructose bisphosphate
30 c3d9sB_
not modelled
9.6
15
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin-5;PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
31 c5y6iB_
not modelled
9.5
26
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator kdgr;PDBTitle: crystal structure of pseudomonas aeruginosa hmgr
32 c4o9uB_
not modelled
9.2
23
PDB header: membrane proteinChain: B: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
33 d1mkma1
not modelled
9.0
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator IclR, N-terminal domain
34 d1a5ca_
not modelled
8.5
29
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
35 c4od5C_
not modelled
7.5
14
PDB header: transferaseChain: C: PDB Molecule: 4-hydroxybenzoate octaprenyltransferase;PDBTitle: substrate-bound structure of a ubia homolog from aeropyrum pernix k1
36 d1f2ja_
not modelled
7.3
33
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
37 c6cfwI_
not modelled
7.1
24
PDB header: membrane proteinChain: I: PDB Molecule: mbh subunit;PDBTitle: cryoem structure of a respiratory membrane-bound hydrogenase
38 d1s7ba_
not modelled
6.9
22
Fold: Multidrug resistance efflux transporter EmrESuperfamily: Multidrug resistance efflux transporter EmrEFamily: Multidrug resistance efflux transporter EmrE
39 c3s0xB_
not modelled
6.2
16
PDB header: hydrolaseChain: B: PDB Molecule: peptidase a24b, flak domain protein;PDBTitle: the crystal structure of gxgd membrane protease flak
40 c2vm6A_
not modelled
6.1
18
PDB header: immune systemChain: A: PDB Molecule: bcl-2-related protein a1;PDBTitle: human bcl2-a1 in complex with bim-bh3 peptide
41 d1u7ga_
not modelled
6.1
21
Fold: Ammonium transporterSuperfamily: Ammonium transporterFamily: Ammonium transporter
42 c5w1eA_
not modelled
6.0
19
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: pobr in complex with phb
43 c5ua4A_
not modelled
5.9
16
PDB header: apoptosisChain: A: PDB Molecule: 5-hl;PDBTitle: crystal structure of a179l:bid bh3 complex
44 c2vofA_
not modelled
5.8
19
PDB header: apoptosisChain: A: PDB Molecule: bcl-2-related protein a1;PDBTitle: structure of mouse a1 bound to the puma bh3-domain
45 c2k1aA_
not modelled
5.8
13
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
46 c4o9tH_
not modelled
5.7
23
PDB header: membrane proteinChain: H: PDB Molecule: nad(p) transhydrogenase subunit beta;PDBTitle: mechanism of transhydrogenase coupling proton translocation and2 hydride transfer
47 d1fdja_
not modelled
5.6
31
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
48 c2pc4B_
not modelled
5.5
29
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose-bisphosphate aldolase from plasmodium2 falciparum in complex with trap-tail determined at 2.4 angstrom3 resolution
49 d2f6ua1
not modelled
5.5
16
Fold: TIM beta/alpha-barrelSuperfamily: FMN-linked oxidoreductasesFamily: FMN-linked oxidoreductases
50 c5aexB_
not modelled
5.5
24
PDB header: membrane proteinChain: B: PDB Molecule: ammonium transporter mep2;PDBTitle: crystal structure of saccharomyces cerevisiae mep2
51 d1v8ba2
not modelled
5.3
17
Fold: Flavodoxin-likeSuperfamily: Formate/glycerate dehydrogenase catalytic domain-likeFamily: S-adenosylhomocystein hydrolase
52 c2d57A_
not modelled
5.3
12
PDB header: transport proteinChain: A: PDB Molecule: aquaporin-4;PDBTitle: double layered 2d crystal structure of aquaporin-4 (aqp4m23) at 3.2 a2 resolution by electron crystallography
53 c6h1nA_
not modelled
5.2
14
PDB header: apoptosisChain: A: PDB Molecule: bcl2-like 10 (apoptosis facilitator);PDBTitle: crystal structure of a zebra-fish pro-survival protein nrz-apo
54 d1xfba1
not modelled
5.1
31
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
55 c4huqS_
not modelled
5.1
16
PDB header: hydrolaseChain: S: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a transporter
56 d1ybea1
not modelled
5.0
27
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: Monomeric nicotinate phosphoribosyltransferase C-terminal domain