1 c4czbB_
100.0
18
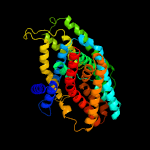
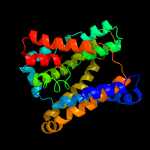
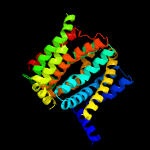
PDB header: membrane proteinChain: B: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: structure of the sodium proton antiporter mjnhap1 from2 methanocaldococcus jannaschii at ph 8.
2 c4cz8A_
100.0
24
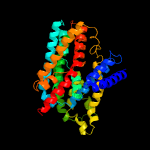
PDB header: membrane proteinChain: A: PDB Molecule: na+/h+ antiporter, putative;PDBTitle: structure of the sodium proton antiporter panhap from2 pyrococcus abyssii at ph 8.
3 c5bz3A_
100.0
18
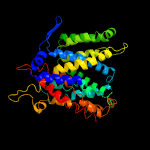
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of sodium proton antiporter napa in outward-facing2 conformation.
4 c4bwzA_
100.0
18
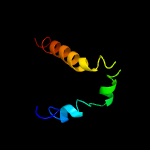
PDB header: transport proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: crystal structure of the sodium proton antiporter, napa
5 c1zcdA_
99.9
11
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter 1;PDBTitle: crystal structure of the na+/h+ antiporter nhaa
6 c2mdfA_
98.8
27
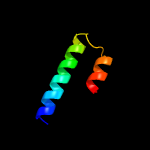
PDB header: proton transportChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: nmr structure of a two-transmembrane segment tm vi-vii of nhe1
7 c4n7wA_
97.5
11
PDB header: transport proteinChain: A: PDB Molecule: transporter, sodium/bile acid symporter family;PDBTitle: crystal structure of the sodium bile acid symporter from yersinia2 frederiksenii
8 c2l0eA_
90.0
21
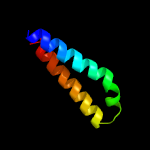
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm vi of the nhe1 isoform of the2 na+/h+ exchanger
9 c2kbvA_
79.0
17
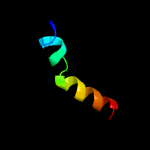
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: structural and functional analysis of tm xi of the nhe12 isoform of the na+/h+ exchanger
10 c6nd1E_
77.2
14
PDB header: protein transportChain: E: PDB Molecule: translocation protein sec66;PDBTitle: cryoem structure of the sec complex from yeast
11 c6b8hb_
75.0
11
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit alpha, mitochondrial;PDBTitle: mosaic model of yeast mitochondrial atp synthase monomer
12 c6j5ib_
59.0
13
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit alpha, mitochondrial;PDBTitle: cryo-em structure of the mammalian dp-state atp synthase
13 c6n3qE_
58.9
17
PDB header: transport proteinChain: E: PDB Molecule: translocation protein sec66;PDBTitle: cryo-em structure of the yeast sec complex
14 c6iu3A_
51.3
13
PDB header: metal transportChain: A: PDB Molecule: vit1;PDBTitle: crystal structure of iron transporter vit1 with zinc ions
15 c4k1cB_
50.1
12
PDB header: membrane protein/metal transportChain: B: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
16 c4k1cA_
48.6
13
PDB header: membrane protein/metal transportChain: A: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
17 d1vqop1
45.7
17
Fold: Ribosomal protein L19 (L19e)Superfamily: Ribosomal protein L19 (L19e)Family: Ribosomal protein L19 (L19e)
18 c5l0wA_
45.4
20
PDB header: membrane proteinChain: A: PDB Molecule: sec71;PDBTitle: structure of post-translational translocation sec71/sec72 complex
19 c2htgA_
41.2
33
PDB header: membrane proteinChain: A: PDB Molecule: nhe1 isoform of na+/h+ exchanger 1;PDBTitle: structural and functional characterization of tm vii of the2 nhe1 isoform of the na+/h+ exchanger
20 c3j21Q_
40.3
19
PDB header: ribosomeChain: Q: PDB Molecule: 50s ribosomal protein l19e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
21 c4a1cO_
not modelled
39.8
14
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
22 c2k3cA_
not modelled
37.9
31
PDB header: metal transportChain: A: PDB Molecule: tmix peptide;PDBTitle: structural and functional characterization of tm ix of the2 nhe1 isoform of the na+/h+ exchanger
23 c4a17O_
not modelled
37.4
14
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
24 d1f6ga_
not modelled
35.4
11
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels
25 c3j39R_
not modelled
34.9
15
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l19;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
26 c2zkrp_
not modelled
34.3
14
PDB header: ribosomal protein/rnaChain: P: PDB Molecule: rna expansion segment es31 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
27 d2csba1
not modelled
32.6
5
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: Topoisomerase V repeat domain
28 c5a1sB_
not modelled
31.5
17
PDB header: transport proteinChain: B: PDB Molecule: citrate-sodium symporter;PDBTitle: crystal structure of the sodium-dependent citrate symporter secits2 form salmonella enterica.
29 c3iz5T_
not modelled
30.7
11
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
30 c2zv4O_
not modelled
29.8
13
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
31 c4b6aR_
not modelled
29.4
20
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l19-b;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
32 c4a1eO_
not modelled
29.0
14
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
33 c3zf7T_
not modelled
28.6
23
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
34 c6fkip_
not modelled
27.9
7
PDB header: membrane proteinChain: P: PDB Molecule: atp synthase subunit c, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
35 c3hzsA_
not modelled
27.9
13
PDB header: transferaseChain: A: PDB Molecule: monofunctional glycosyltransferase;PDBTitle: s. aureus monofunctional glycosyltransferase (mtga)in complex with2 moenomycin
36 c6b3rE_
not modelled
27.7
5
PDB header: transport proteinChain: E: PDB Molecule: piezo-type mechanosensitive ion channel component 1;PDBTitle: structure of the mechanosensitive channel piezo1
37 d2nwwa1
not modelled
27.1
14
Fold: Proton glutamate symport proteinSuperfamily: Proton glutamate symport proteinFamily: Proton glutamate symport protein
38 c2m7xA_
not modelled
25.1
11
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: structural and functional analysis of transmembrane segment iv of the2 salt tolerance protein sod2
39 c6epgD_
not modelled
25.0
13
PDB header: toxinChain: D: PDB Molecule: zeta_1 toxin;PDBTitle: structure of the epsilon_1 / zeta_1 antitoxin / toxin system from2 neisseria gonorrhoeae.
40 c3f0cA_
not modelled
24.3
6
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of transcriptional regulator from cytophaga2 hutchinsonii atcc 33406
41 c5nugB_
not modelled
20.3
8
PDB header: motor proteinChain: B: PDB Molecule: cytoplasmic dynein 1 heavy chain 1;PDBTitle: motor domains from human cytoplasmic dynein-1 in the phi-particle2 conformation
42 c5arhT_
not modelled
20.2
14
PDB header: hydrolaseChain: T: PDB Molecule: atp synthase f(0) complex subunit b1, mitochondrial;PDBTitle: bovine mitochondrial atp synthase state 2a
43 c6eznH_
not modelled
19.6
16
PDB header: membrane proteinChain: H: PDB Molecule: dolichyl-diphosphooligosaccharide--proteinPDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
44 c4l6rA_
not modelled
19.2
16
PDB header: membrane proteinChain: A: PDB Molecule: soluble cytochrome b562 and glucagon receptor chimera;PDBTitle: structure of the class b human glucagon g protein coupled receptor
45 c6iiuA_
not modelled
19.1
19
PDB header: signaling proteinChain: A: PDB Molecule: soluble cytochrome b562,thromboxane a2 receptor,rubredoxin,PDBTitle: crystal structure of the human thromboxane a2 receptor bound to2 ramatroban
46 c1y4cA_
not modelled
17.1
9
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
47 c5lqyV_
not modelled
16.8
14
PDB header: hydrolaseChain: V: PDB Molecule: atp synthase subunit b;PDBTitle: structure of f-atpase from pichia angusta, in state2
48 d2bfda1
not modelled
16.8
7
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase PP module
49 c3aqpB_
not modelled
16.7
21
PDB header: membrane proteinChain: B: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
50 c5z96C_
not modelled
16.6
11
PDB header: membrane proteinChain: C: PDB Molecule: short transient receptor potential channel 4;PDBTitle: structure of the mouse trpc4 ion channel
51 d1umda_
not modelled
16.3
6
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase PP module
52 c5mqfQ_
not modelled
15.2
9
PDB header: splicingChain: Q: PDB Molecule: protein bud31 homolog;PDBTitle: cryo-em structure of a human spliceosome activated for step 2 of2 splicing (c* complex)
53 c5xamA_
not modelled
14.6
18
PDB header: membrane proteinChain: A: PDB Molecule: protein translocase subunit secd;PDBTitle: crystal structure of secdf in i form at 4 a resolution
54 c2yicC_
not modelled
14.4
16
PDB header: lyaseChain: C: PDB Molecule: 2-oxoglutarate decarboxylase;PDBTitle: crystal structure of the suca domain of mycobacterium smegmatis2 alpha-ketoglutarate decarboxylase (triclinic form)
55 c3vuqD_
not modelled
13.2
10
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (tetr/acrr family);PDBTitle: crystal structure of ttha0167, a transcriptional regulator, tetr/acrr2 family from thermus thermophilus hb8
56 c4gctA_
not modelled
13.1
8
PDB header: dna binding protein/dnaChain: A: PDB Molecule: nucleoid occlusion factor slma;PDBTitle: structure of no factor protein-dna complex
57 c4q28B_
not modelled
12.5
25
PDB header: structural proteinChain: B: PDB Molecule: periplakin;PDBTitle: crystal structure of the plectin 1 and 2 repeats of the human2 periplakin. northeast structural genomics consortium (nesg) target3 hr9083a
58 c3aygA_
not modelled
12.0
6
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric oxide reductase;PDBTitle: crystal structure of nitric oxide reductase complex with hqno
59 c5t4oJ_
not modelled
11.9
13
PDB header: hydrolaseChain: J: PDB Molecule: atp synthase subunit b;PDBTitle: autoinhibited e. coli atp synthase state 1
60 c3exmA_
not modelled
11.8
8
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase sc4828;PDBTitle: crystal structure of the phosphatase sc4828 with the non-hydrolyzable2 nucleotide gpcp
61 c3thzA_
not modelled
11.7
12
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna mismatch repair protein msh2;PDBTitle: human mutsbeta complexed with an idl of 6 bases (loop6) and adp
62 c6flnE_
not modelled
11.6
11
PDB header: protein bindingChain: E: PDB Molecule: e3 ubiquitin/isg15 ligase trim25;PDBTitle: crystal structure of the human trim25 coiled-coil and pryspry domains
63 c6c9aB_
not modelled
11.6
12
PDB header: membrane proteinChain: B: PDB Molecule: two pore calcium channel protein 1;PDBTitle: cryo-em structure of mouse tpc1 channel in the ptdins(3,5)p2-bound2 state
64 c4jkvA_
not modelled
11.2
16
PDB header: membrane proteinChain: A: PDB Molecule: soluble cytochrome b562, smoothened homolog;PDBTitle: structure of the human smoothened 7tm receptor in complex with an2 antitumor agent
65 c4k0eA_
not modelled
10.7
13
PDB header: transport proteinChain: A: PDB Molecule: heavy metal cation tricomponent efflux pump znea(czca-PDBTitle: x-ray crystal structure of a heavy metal efflux pump, crystal form ii
66 c4cg4D_
not modelled
10.7
7
PDB header: actin-binding proteinChain: D: PDB Molecule: pyrin;PDBTitle: crystal structure of the chs-b30.2 domains of trim20
67 c1y4eA_
not modelled
10.4
24
PDB header: membrane proteinChain: A: PDB Molecule: sodium/hydrogen exchanger 1;PDBTitle: nmr structure of transmembrane segment iv of the nhe12 isoform of the na+/h+ exchanger
68 c2i10A_
not modelled
10.3
10
PDB header: transcriptionChain: A: PDB Molecule: putative tetr transcriptional regulator;PDBTitle: putative tetr transcriptional regulator from rhodococcus sp. rha1
69 d2oeza1
not modelled
10.2
24
Fold: YacF-likeSuperfamily: YacF-likeFamily: YacF-like
70 c4n6hA_
not modelled
9.9
16
PDB header: signaling proteinChain: A: PDB Molecule: soluble cytochrome b562, delta-type opioid receptorPDBTitle: 1.8 a structure of the human delta opioid 7tm receptor (psi community2 target)
71 d2c0sa1
not modelled
9.7
6
Fold: ROP-likeSuperfamily: BAS1536-likeFamily: BAS1536-like
72 c5c4iC_
not modelled
9.2
18
PDB header: oxidoreductaseChain: C: PDB Molecule: oxalate oxidoreductase subunit beta;PDBTitle: structure of an oxalate oxidoreductase
73 c6cnmA_
not modelled
9.1
8
PDB header: membrane proteinChain: A: PDB Molecule: intermediate conductance calcium-activated potassiumPDBTitle: cryo-em structure of the human sk4/calmodulin channel complex
74 c5zdnA_
not modelled
9.1
3
PDB header: hydrolaseChain: A: PDB Molecule: fomd;PDBTitle: the complex structure of fomd with cdp
75 c4dncD_
not modelled
9.0
29
PDB header: transcriptionChain: D: PDB Molecule: male-specific lethal 1 homolog;PDBTitle: crystal structure of human mof in complex with msl1
76 c2lx0A_
not modelled
9.0
15
PDB header: membrane proteinChain: A: PDB Molecule: membrane fusion protein p14;PDBTitle: arced helix (arch) nmr structure of the reovirus p14 fusion-associated2 small transmembrane (fast) protein transmembrane domain (tmd) in3 dodecyl phosphocholine (dpc) micelles
77 c6fkib_
not modelled
8.9
12
PDB header: membrane proteinChain: B: PDB Molecule: atp synthase subunit beta, chloroplastic;PDBTitle: chloroplast f1fo conformation 3
78 d2oqoa1
not modelled
8.6
13
Fold: Lysozyme-likeSuperfamily: Lysozyme-likeFamily: PBP transglycosylase domain-like
79 c6b87C_
not modelled
8.5
23
PDB header: membrane proteinChain: C: PDB Molecule: tmhc2_e;PDBTitle: crystal structure of transmembrane protein tmhc2_e
80 c3kltB_
not modelled
8.5
18
PDB header: structural proteinChain: B: PDB Molecule: vimentin;PDBTitle: crystal structure of a vimentin fragment
81 c2my1A_
not modelled
8.4
13
PDB header: splicingChain: A: PDB Molecule: pre-mrna-splicing factor bud31;PDBTitle: solution structure of bud31p
82 c2g7sA_
not modelled
8.4
6
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, tetr family;PDBTitle: the crystal structure of transcriptional regulator, tetr family, from2 agrobacterium tumefaciens
83 c5eulE_
not modelled
8.4
17
PDB header: protein transportChain: E: PDB Molecule: preprotein translocase sece subunit;PDBTitle: structure of the seca-secy complex with a translocating polypeptide2 substrate
84 c3vmtA_
not modelled
8.4
13
PDB header: transferaseChain: A: PDB Molecule: monofunctional glycosyltransferase;PDBTitle: crystal structure of staphylococcus aureus membrane-bound2 transglycosylase in complex with a lipid ii analog
85 c2l2lA_
not modelled
8.3
28
PDB header: transferaseChain: A: PDB Molecule: transcriptional repressor p66-alpha;PDBTitle: solution structure of the coiled-coil complex between mbd2 and2 p66alpha
86 c6qajB_
not modelled
8.3
10
PDB header: nuclear proteinChain: B: PDB Molecule: endolysin,transcription intermediary factor 1-beta;PDBTitle: structure of the tripartite motif of kap1/trim28
87 c5dkoA_
not modelled
8.2
18
PDB header: replicationChain: A: PDB Molecule: cell division protein zapd;PDBTitle: the structure of escherichia coli zapd
88 d1w85a_
not modelled
8.2
5
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase PP module
89 c3t57A_
not modelled
8.1
11
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine o-acyltransferase domain-containingPDBTitle: activity and crystal structure of arabidopsis udp-n-acetylglucosamine2 acyltransferase
90 c6ayiA_
not modelled
8.1
8
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional regulator uidr;PDBTitle: escherichia coli gusr
91 c5j3nA_
not modelled
8.0
11
PDB header: hydrolaseChain: A: PDB Molecule: green fluorescent protein,hsdr;PDBTitle: c-terminal domain of ecor124i hsdr subunit fused with the ph-sensitive2 gfp variant ratiometric phluorin
92 d1zk8a2
not modelled
8.0
10
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
93 c5cbfA_
not modelled
7.8
14
PDB header: transport proteinChain: A: PDB Molecule: ion transport 2 domain protein;PDBTitle: structural and functional characterization of a calcium-activated2 cation channel from tsukamurella paurometabola
94 c1vi0B_
not modelled
7.8
10
PDB header: transcriptionChain: B: PDB Molecule: transcriptional regulator;PDBTitle: crystal structure of a transcriptional regulator
95 c3jb9e_
not modelled
7.7
16
PDB header: rna binding protein/rnaChain: E: PDB Molecule: small nuclear ribonucleoprotein-associated protein b;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
96 c1yo7A_
not modelled
7.7
11
PDB header: replication regulatorChain: A: PDB Molecule: regulatory protein rop;PDBTitle: re-engineering topology of the homodimeric rop protein into a single-2 chain 4-helix bundle
97 d1i0aa_
not modelled
7.7
9
Fold: L-aspartase-likeSuperfamily: L-aspartase-likeFamily: L-aspartase/fumarase
98 c5hg1A_
not modelled
7.7
22
PDB header: transferase/transferase inhibitorChain: A: PDB Molecule: hexokinase-2;PDBTitle: crystal structure of human hexokinase 2 with cmpd 1, a c-2-substituted2 glucosamine
99 c5frgA_
not modelled
7.6
10
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1