| 1 |
|
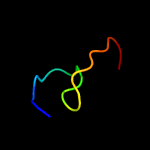
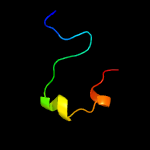
PDB 2elr chain A
Region: 13 - 31
Aligned: 19
Modelled: 19
Confidence: 29.9%
Identity: 37%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 15th c2h2 zinc finger of human2 zinc finger protein 406
Phyre2
| 2 |
|
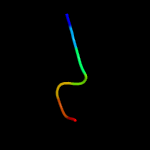
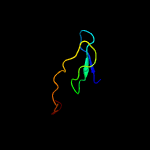
PDB 1tx6 chain J
Region: 57 - 69
Aligned: 13
Modelled: 13
Confidence: 11.9%
Identity: 62%
PDB header:hydrolase/protein binding
Chain: J: PDB Molecule:bowman-birk type trypsin inhibitor;
PDBTitle: trypsin:bbi complex
Phyre2
| 3 |
|
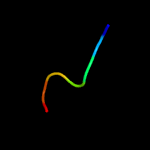
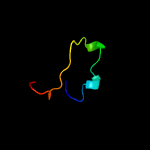
PDB 3c97 chain A
Region: 51 - 65
Aligned: 15
Modelled: 15
Confidence: 8.4%
Identity: 27%
PDB header:signaling protein, transferase
Chain: A: PDB Molecule:signal transduction histidine kinase;
PDBTitle: crystal structure of the response regulator receiver domain of a2 signal transduction histidine kinase from aspergillus oryzae
Phyre2
| 4 |
|
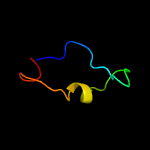
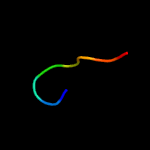
PDB 3jtc chain C
Region: 17 - 23
Aligned: 7
Modelled: 7
Confidence: 8.4%
Identity: 57%
PDB header:blood clotting
Chain: C: PDB Molecule:vitamin k-dependent protein c;
PDBTitle: importance of mg2+ in the ca2+-dependent folding of the gamma-2 carboxyglutamic acid domains of vitamin k-dependent clotting and3 anticlotting proteins
Phyre2
| 5 |
|
PDB 3jtc chain D
Region: 17 - 23
Aligned: 7
Modelled: 7
Confidence: 8.4%
Identity: 57%
PDB header:blood clotting
Chain: D: PDB Molecule:vitamin k-dependent protein c;
PDBTitle: importance of mg2+ in the ca2+-dependent folding of the gamma-2 carboxyglutamic acid domains of vitamin k-dependent clotting and3 anticlotting proteins
Phyre2
| 6 |
|
PDB 1lqv chain C
Region: 17 - 23
Aligned: 7
Modelled: 7
Confidence: 8.4%
Identity: 57%
PDB header:blood clotting
Chain: C: PDB Molecule:vitamin-k dependent protein c;
PDBTitle: crystal structure of the endothelial protein c receptor with2 phospholipid in the groove in complex with gla domain of protein c.
Phyre2
| 7 |
|
PDB 1lqv chain D
Region: 17 - 23
Aligned: 7
Modelled: 7
Confidence: 8.4%
Identity: 57%
PDB header:blood clotting
Chain: D: PDB Molecule:vitamin-k dependent protein c;
PDBTitle: crystal structure of the endothelial protein c receptor with2 phospholipid in the groove in complex with gla domain of protein c.
Phyre2
| 8 |
|
PDB 3g3l chain A
Region: 21 - 55
Aligned: 32
Modelled: 32
Confidence: 8.0%
Identity: 47%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative uncharacterized membrane-associated protein;
PDBTitle: crystal structure of putative membrane-associated protein of unknown2 function (yp_211325.1) from bacteroides fragilis nctc 9343 at 2.20 a3 resolution
Phyre2
| 9 |
|
PDB 6re8 chain 0
Region: 35 - 62
Aligned: 28
Modelled: 28
Confidence: 7.9%
Identity: 32%
PDB header:proton transport
Chain: 0: PDB Molecule:asa-10: polytomella f-atp synthase associated subunit 10;
PDBTitle: cryo-em structure of polytomella f-atp synthase, rotary substate 2d,2 composite map
Phyre2
| 10 |
|
PDB 3a9k chain C
Region: 11 - 23
Aligned: 13
Modelled: 13
Confidence: 7.2%
Identity: 38%
PDB header:signaling protein/metal binding protein
Chain: C: PDB Molecule:mitogen-activated protein kinase kinase kinase 7-
PDBTitle: crystal structure of the mouse tab3-nzf in complex with2 lys63-linked di-ubiquitin
Phyre2
| 11 |
|
PDB 2i7f chain B
Region: 16 - 55
Aligned: 40
Modelled: 40
Confidence: 7.0%
Identity: 15%
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin component of dioxygenase;
PDBTitle: sphingomonas yanoikuyae b1 ferredoxin
Phyre2
| 12 |
|
PDB 4o1i chain A
Region: 41 - 66
Aligned: 26
Modelled: 26
Confidence: 6.4%
Identity: 31%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulatory protein;
PDBTitle: crystal structure of the regulatory domain of mtbglnr
Phyre2
| 13 |
|
PDB 2ayx chain A
Region: 51 - 65
Aligned: 15
Modelled: 15
Confidence: 6.0%
Identity: 33%
PDB header:transferase
Chain: A: PDB Molecule:sensor kinase protein rcsc;
PDBTitle: solution structure of the e.coli rcsc c-terminus (residues2 700-949) containing linker region and phosphoreceiver3 domain
Phyre2
| 14 |
|
PDB 6dzp chain Y
Region: 21 - 28
Aligned: 8
Modelled: 8
Confidence: 5.4%
Identity: 38%
PDB header:ribosome
Chain: Y: PDB Molecule:
PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 50s ribosomal2 subunit
Phyre2
| 15 |
|
PDB 2js3 chain B
Region: 9 - 21
Aligned: 13
Modelled: 13
Confidence: 5.3%
Identity: 54%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: nmr structure of protein q6n9a4_rhopa. northeast structural genomics2 consortium target rpt8
Phyre2
| 16 |
|
PDB 2ae8 chain A domain 2
Region: 12 - 56
Aligned: 32
Modelled: 32
Confidence: 5.2%
Identity: 41%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: Imidazole glycerol phosphate dehydratase
Phyre2
| 17 |
|
PDB 1m9s chain A domain 2
Region: 12 - 37
Aligned: 26
Modelled: 26
Confidence: 5.1%
Identity: 31%
Fold: SH3-like barrel
Superfamily: Prokaryotic SH3-related domain
Family: GW domain
Phyre2
| 18 |
|
PDB 6ddg chain J
Region: 21 - 28
Aligned: 8
Modelled: 8
Confidence: 5.0%
Identity: 38%
PDB header:ribosome/antibiotic
Chain: J: PDB Molecule:50s ribosomal protein l28;
PDBTitle: structure of the 50s ribosomal subunit from methicillin resistant2 staphylococcus aureus in complex with the oxazolidinone antibiotic3 lzd-6
Phyre2
| 19 |
|
PDB 2qn5 chain B
Region: 57 - 68
Aligned: 12
Modelled: 12
Confidence: 5.0%
Identity: 33%
PDB header:hydrolase inhibitor/hydrolase
Chain: B: PDB Molecule:bowman-birk type bran trypsin inhibitor;
PDBTitle: crystal structure and functional study of the bowman-birk2 inhibitor from rice bran in complex with bovine trypsin
Phyre2