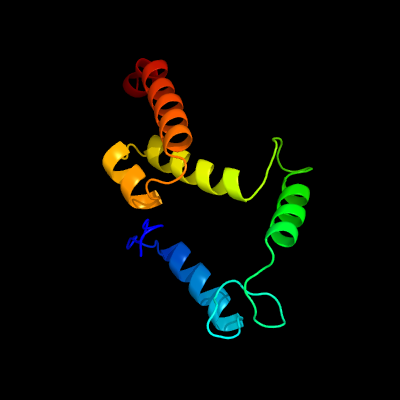
1 c3nkhB_
99.9
23
PDB header: recombinationChain: B: PDB Molecule: integrase;PDBTitle: crystal structure of integrase from mrsa strain staphylococcus aureus
2 c5jjvA_
99.9
23
PDB header: recombinationChain: A: PDB Molecule: tyrosine recombinase xerh;PDBTitle: crystal structure of xerh site-specific recombinase bound to2 palindromic difh substrate: post-cleavage complex
3 c6en2A_
99.9
25
PDB header: recombinationChain: A: PDB Molecule: int protein;PDBTitle: structure of the tn1549 transposon integrase (aa 82-397, r225k) in2 complex with a circular intermediate dna (ci6b-dna)
4 c5dcfA_
99.9
29
PDB header: recombinationChain: A: PDB Molecule: tyrosine recombinase xerd,dna translocase ftsk;PDBTitle: c-terminal domain of xerd recombinase in complex with gamma domain of2 ftsk
5 c5vfzA_
99.9
20
PDB header: dna binding proteinChain: A: PDB Molecule: gp33;PDBTitle: integrase from mycobacterium phage brujita
6 c2a3vA_
99.8
26
PDB header: recombinationChain: A: PDB Molecule: site-specific recombinase inti4;PDBTitle: structural basis for broad dna-specificity in integron recombination
7 c5hxyE_
99.8
26
PDB header: recombinationChain: E: PDB Molecule: tyrosine recombinase xera;PDBTitle: crystal structure of xera recombinase
8 c1ma7A_
99.8
12
PDB header: hydrolase, ligase/dnaChain: A: PDB Molecule: cre recombinase;PDBTitle: crystal structure of cre site-specific recombinase2 complexed with a mutant dna substrate, loxp-a8/t27
9 d1aiha_
99.8
24
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
10 c1z1bA_
99.7
23
PDB header: dna binding protein/dnaChain: A: PDB Molecule: integrase;PDBTitle: crystal structure of a lambda integrase dimer bound to a2 coc' core site
11 c1crxA_
99.7
13
PDB header: replication/dnaChain: A: PDB Molecule: cre recombinase;PDBTitle: cre recombinase/dna complex reaction intermediate i
12 c5c6kB_
99.7
24
PDB header: hydrolaseChain: B: PDB Molecule: integrase;PDBTitle: bacteriophage p2 integrase catalytic domain
13 c1a0pA_
99.7
23
PDB header: dna recombinationChain: A: PDB Molecule: site-specific recombinase xerd;PDBTitle: site-specific recombinase, xerd
14 c4a8eA_
99.6
21
PDB header: cell cycleChain: A: PDB Molecule: probable tyrosine recombinase xerc-like;PDBTitle: the structure of a dimeric xer recombinase from archaea
15 d1p7da_
99.6
21
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
16 c3uxuA_
99.6
13
PDB header: recombinationChain: A: PDB Molecule: probable integrase;PDBTitle: the structure of the catalytic domain of the sulfolobus spindle-shaped2 viral integrase reveals an evolutionarily conserved catalytic core3 and supports a mechanism of dna cleavage in trans
17 d1f44a2
99.5
15
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
18 d1a0pa2
99.5
28
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
19 d1ae9a_
99.4
20
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
20 d5crxb2
99.0
13
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Lambda integrase-like, catalytic core
21 c2f4qA_
not modelled
92.4
17
PDB header: isomeraseChain: A: PDB Molecule: type i topoisomerase, putative;PDBTitle: crystal structure of deinococcus radiodurans topoisomerase ib
22 c2v6eB_
not modelled
89.0
13
PDB header: hydrolaseChain: B: PDB Molecule: protelemorase;PDBTitle: protelomerase telk complexed with substrate dna
23 c4f43A_
not modelled
88.9
17
PDB header: recombination/dnaChain: A: PDB Molecule: protelomerase;PDBTitle: protelomerase tela mutant r255a complexed with caag hairpin dna
24 c2h7fX_
not modelled
87.3
14
PDB header: isomerase/dnaChain: X: PDB Molecule: dna topoisomerase 1;PDBTitle: structure of variola topoisomerase covalently bound to dna
25 c2b9sA_
not modelled
76.4
18
PDB header: isomerase/dnaChain: A: PDB Molecule: topoisomerase i-like protein;PDBTitle: crystal structure of heterodimeric l. donovani topoisomerase i-2 vanadate-dna complex
26 d1a41a_
not modelled
75.3
14
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Eukaryotic DNA topoisomerase I, catalytic core
27 d1k4ta2
not modelled
74.8
20
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Eukaryotic DNA topoisomerase I, catalytic core
28 d1rr8c1
not modelled
63.4
20
Fold: DNA breaking-rejoining enzymesSuperfamily: DNA breaking-rejoining enzymesFamily: Eukaryotic DNA topoisomerase I, catalytic core
29 c1rrjA_
not modelled
58.7
20
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase i;PDBTitle: structural mechanisms of camptothecin resistance by2 mutations in human topoisomerase i
30 c1a31A_
not modelled
53.6
20
PDB header: isomerase/dnaChain: A: PDB Molecule: protein (topoisomerase i);PDBTitle: human reconstituted dna topoisomerase i in covalent complex2 with a 22 base pair dna duplex
31 d1l8qa1
not modelled
39.8
11
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Chromosomal replication initiation factor DnaA C-terminal domain IV
32 d6paxa1
not modelled
22.8
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
33 d1j1va_
not modelled
20.0
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: Chromosomal replication initiation factor DnaA C-terminal domain IV
34 c2mqkA_
not modelled
18.7
13
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent target dna activator b;PDBTitle: solution structure of n terminal domain of the mub aaa+ atpase
35 c3sohB_
not modelled
16.5
14
PDB header: motor proteinChain: B: PDB Molecule: flagellar motor switch protein flig;PDBTitle: architecture of the flagellar rotor
36 c1p4eB_
not modelled
15.3
18
PDB header: dna binding protein/recombination/dnaChain: B: PDB Molecule: recombinase flp protein;PDBTitle: flpe w330f mutant-dna holliday junction complex
37 c1nh3A_
not modelled
14.3
20
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase i;PDBTitle: human topoisomerase i ara-c complex
38 d1k78a1
not modelled
13.6
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain
39 c2uwqA_
not modelled
13.1
15
PDB header: apoptosisChain: A: PDB Molecule: apoptosis-stimulating of p53 protein 2;PDBTitle: solution structure of aspp2 n-terminus
40 c5zktA_
not modelled
12.2
17
PDB header: transcriptionChain: A: PDB Molecule: putative transcription factor pcf6;PDBTitle: crystal structure of tcp domain of pcf6 in oryza sativa
41 c3oouA_
not modelled
10.5
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: lin2118 protein;PDBTitle: the structure of a protein with unkown function from listeria innocua
42 c5zktB_
not modelled
9.9
17
PDB header: transcriptionChain: B: PDB Molecule: putative transcription factor pcf6;PDBTitle: crystal structure of tcp domain of pcf6 in oryza sativa
43 d1g99a1
not modelled
8.3
24
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like
44 c4m70E_
not modelled
8.0
16
PDB header: plant proteinChain: E: PDB Molecule: ran gtpase activating protein 2;PDBTitle: crystal structure of potato rx-cc domain in complex with rangap2-wpp2 domain
45 c3pvpA_
not modelled
8.0
11
PDB header: dna binding protein/dnaChain: A: PDB Molecule: chromosomal replication initiator protein dnaa;PDBTitle: structure of mycobacterium tuberculosis dnaa-dbd in complex with box22 dna
46 d2vlqa1
not modelled
7.7
11
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins
47 d1gxha_
not modelled
7.2
11
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins
48 d1ayia_
not modelled
6.3
11
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins
49 d1xc5a1
not modelled
6.3
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain
50 c2v1lA_
not modelled
6.3
16
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of the conserved hypothetical protein vc1805 from2 pathogenicity island vpi-2 of vibrio cholerae o1 biovar3 eltor str. n16961 shares structural homology with the4 human p32 protein
51 c4qkoA_
not modelled
5.7
5
PDB header: antimicrobial proteinChain: A: PDB Molecule: pyocin-s2 immunity protein;PDBTitle: the crystal structure of the pyocin s2 nuclease domain, immunity2 protein complex at 1.8 angstroms
52 c3lsgD_
not modelled
5.7
9
PDB header: transcription regulatorChain: D: PDB Molecule: two-component response regulator yesn;PDBTitle: the crystal structure of the c-terminal domain of the two-2 component response regulator yesn from fusobacterium3 nucleatum subsp. nucleatum atcc 25586
53 d2e1za1
not modelled
5.4
22
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Acetokinase-like
54 c3pkrA_
not modelled
5.4
7
PDB header: motor proteinChain: A: PDB Molecule: flagellar motor switch protein;PDBTitle: crystal structure of flig (residue 86-343) from h. pylori
55 c2eqrA_
not modelled
5.4
10
PDB header: transcriptionChain: A: PDB Molecule: nuclear receptor corepressor 1;PDBTitle: solution structure of the first sant domain from human2 nuclear receptor corepressor 1
56 c2jwlB_
not modelled
5.3
22
PDB header: membrane proteinChain: B: PDB Molecule: protein tolr;PDBTitle: solution structure of periplasmic domain of tolr from h.2 influenzae with saxs data
57 d2auwa1
not modelled
5.2
7
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like
58 c6ioyD_
not modelled
5.2
25
PDB header: transferaseChain: D: PDB Molecule: acetate kinase;PDBTitle: crystal structure of porphyromonas gingivalis acetate kinase
59 c4a69C_
not modelled
5.1
13
PDB header: transcriptionChain: C: PDB Molecule: nuclear receptor corepressor 2;PDBTitle: structure of hdac3 bound to corepressor and inositol tetraphosphate
60 c2no8A_
not modelled
5.1
12
PDB header: immune systemChain: A: PDB Molecule: colicin-e2 immunity protein;PDBTitle: nmr structure analysis of the colicin immuntiy protein im2