| 1 |
|
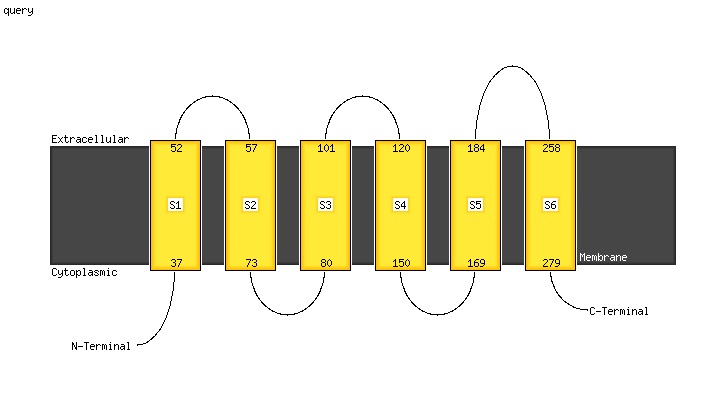
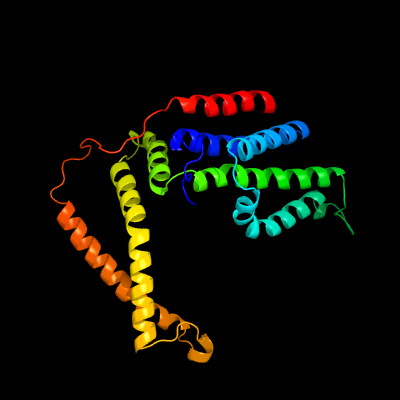
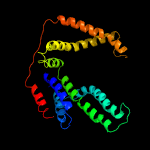
PDB 5d7t chain H
Region: 24 - 273
Aligned: 247
Modelled: 250
Confidence: 100.0%
Identity: 18%
PDB header:transport protein
Chain: H: PDB Molecule:energy-coupling factor transporter transmembrane protein
PDBTitle: folate ecf transporter: apo state
Phyre2
| 2 |
|
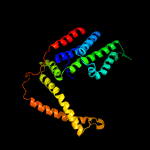
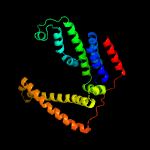
PDB 4huq chain T
Region: 24 - 274
Aligned: 239
Modelled: 251
Confidence: 100.0%
Identity: 21%
PDB header:hydrolase
Chain: T: PDB Molecule:energy-coupling factor transporter transmembrane protein
PDBTitle: crystal structure of a transporter
Phyre2
| 3 |
|
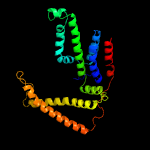
PDB 4rfs chain T
Region: 32 - 267
Aligned: 231
Modelled: 232
Confidence: 100.0%
Identity: 19%
PDB header:hydrolase, transport protein
Chain: T: PDB Molecule:energy-coupling factor transporter transmembrane protein
PDBTitle: structure of a pantothenate energy coupling factor transporter
Phyre2
| 4 |
|
PDB 5x3x chain Q
Region: 28 - 273
Aligned: 227
Modelled: 246
Confidence: 100.0%
Identity: 20%
PDB header:transport protein
Chain: Q: PDB Molecule:uncharacterized protein cbiq;
PDBTitle: 2.8a resolution structure of a cobalt energy-coupling factor2 transporter-cbimqo
Phyre2
| 5 |
|
PDB 3hd7 chain A
Region: 229 - 282
Aligned: 52
Modelled: 54
Confidence: 17.9%
Identity: 12%
PDB header:exocytosis
Chain: A: PDB Molecule:vesicle-associated membrane protein 2;
PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
Phyre2
| 6 |
|
PDB 2xzm chain L
Region: 24 - 36
Aligned: 13
Modelled: 13
Confidence: 17.5%
Identity: 23%
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s12;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
Phyre2
| 7 |
|
PDB 1s1h chain L
Region: 24 - 36
Aligned: 13
Modelled: 13
Confidence: 17.3%
Identity: 15%
PDB header:ribosome
Chain: L: PDB Molecule:40s ribosomal protein s23;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
Phyre2
| 8 |
|
PDB 3u5g chain X
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 14.6%
Identity: 17%
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein s23-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
Phyre2
| 9 |
|
PDB 1k4t chain A domain 2
Region: 10 - 43
Aligned: 30
Modelled: 34
Confidence: 13.5%
Identity: 20%
Fold: DNA breaking-rejoining enzymes
Superfamily: DNA breaking-rejoining enzymes
Family: Eukaryotic DNA topoisomerase I, catalytic core
Phyre2
| 10 |
|
PDB 3j20 chain N
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 12.9%
Identity: 25%
PDB header:ribosome
Chain: N: PDB Molecule:30s ribosomal protein s12p;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
Phyre2
| 11 |
|
PDB 2zkq chain L
Region: 24 - 36
Aligned: 13
Modelled: 13
Confidence: 12.2%
Identity: 15%
PDB header:ribosomal protein/rna
Chain: L: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
Phyre2
| 12 |
|
PDB 3zey chain S
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 10.8%
Identity: 25%
PDB header:ribosome
Chain: S: PDB Molecule:40s ribosomal protein s23, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
Phyre2
| 13 |
|
PDB 2mv6 chain A
Region: 16 - 53
Aligned: 31
Modelled: 38
Confidence: 10.3%
Identity: 29%
PDB header:membrane protein
Chain: A: PDB Molecule:erythropoietin receptor;
PDBTitle: solution structure of the transmembrane domain and the juxta-membrane2 domain of the erythropoietin receptor in micelles
Phyre2
| 14 |
|
PDB 5xyi chain X
Region: 24 - 35
Aligned: 12
Modelled: 12
Confidence: 8.7%
Identity: 17%
PDB header:ribosome
Chain: X: PDB Molecule:40s ribosomal protein s23, putative;
PDBTitle: small subunit of trichomonas vaginalis ribosome
Phyre2
| 15 |
|
PDB 5jx6 chain C
Region: 195 - 259
Aligned: 50
Modelled: 50
Confidence: 6.1%
Identity: 20%
PDB header:hydrolase
Chain: C: PDB Molecule:glucanase;
PDBTitle: gh6 orpinomyces sp. y102 enzyme
Phyre2
| 16 |
|
PDB 3skq chain A
Region: 188 - 219
Aligned: 32
Modelled: 32
Confidence: 5.9%
Identity: 19%
PDB header:metal transport
Chain: A: PDB Molecule:mitochondrial distribution and morphology protein 38;
PDBTitle: mdm38 is a 14-3-3-like receptor and associates with the protein2 synthesis machinery at the inner mitochondrial membrane
Phyre2
| 17 |
|
PDB 2lsm chain A
Region: 227 - 246
Aligned: 20
Modelled: 20
Confidence: 5.7%
Identity: 20%
PDB header:chaperone
Chain: A: PDB Molecule:dna-packaging protein fi;
PDBTitle: solution structure of gpfi c-terminal domain
Phyre2
| 18 |
|
PDB 3ol0 chain C
Region: 5 - 26
Aligned: 22
Modelled: 22
Confidence: 5.3%
Identity: 27%
PDB header:de novo protein
Chain: C: PDB Molecule:de novo designed monomer trefoil-fold sub-domain which
PDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
Phyre2