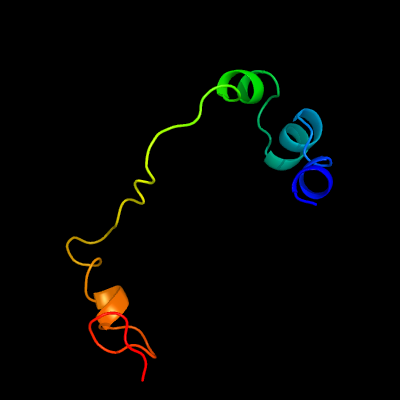
1 d1kqfa2
99.2
29
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
2 c1kqgA_
99.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: formate dehydrogenase, nitrate-inducible, major subunit;PDBTitle: formate dehydrogenase n from e. coli
3 c1h0hA_
98.8
27
PDB header: electron transportChain: A: PDB Molecule: formate dehydrogenase subunit alpha;PDBTitle: tungsten containing formate dehydrogenase from desulfovibrio gigas
4 d1h0ha2
98.8
25
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
5 d1ogya2
98.8
29
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
6 d2jioa2
98.7
35
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
7 c4aayE_
98.6
27
PDB header: oxidoreductaseChain: E: PDB Molecule: aroa;PDBTitle: crystal structure of the arsenite oxidase protein complex2 from rhizobium species strain nt-26
8 c2nyaF_
98.5
30
PDB header: oxidoreductaseChain: F: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the periplasmic nitrate reductase2 (nap) from escherichia coli
9 c2v45A_
98.5
36
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: a new catalytic mechanism of periplasmic nitrate reductase2 from desulfovibrio desulfuricans atcc 27774 from3 crystallographic and epr data and based on detailed4 analysis of the sixth ligand
10 d2iv2x2
98.4
29
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
11 c2iv2X_
98.3
29
PDB header: oxidoreductaseChain: X: PDB Molecule: formate dehydrogenase h;PDBTitle: reinterpretation of reduced form of formate dehydrogenase h from e.2 coli
12 c1ogyA_
98.3
29
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic nitrate reductase;PDBTitle: crystal structure of the heterodimeric nitrate reductase2 from rhodobacter sphaeroides
13 d1y5ia2
98.2
23
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
14 c2ivfA_
98.2
14
PDB header: oxidoreductaseChain: A: PDB Molecule: ethylbenzene dehydrogenase alpha-subunit;PDBTitle: ethylbenzene dehydrogenase from aromatoleum aromaticum
15 c5ch7E_
98.1
25
PDB header: oxidoreductaseChain: E: PDB Molecule: dmso reductase family type ii enzyme, molybdopterinPDBTitle: crystal structure of the perchlorate reductase pcrab - phe164 gate2 switch intermediate - from azospira suillum ps
16 d1g8ka2
97.8
34
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
17 c1g8jC_
97.8
30
PDB header: oxidoreductaseChain: C: PDB Molecule: arsenite oxidase;PDBTitle: crystal structure analysis of arsenite oxidase from2 alcaligenes faecalis
18 c2vpyE_
97.8
14
PDB header: oxidoreductaseChain: E: PDB Molecule: thiosulfate reductase;PDBTitle: polysulfide reductase with bound quinone inhibitor,2 pentachlorophenol (pcp)
19 c1y5iA_
97.8
23
PDB header: oxidoreductaseChain: A: PDB Molecule: respiratory nitrate reductase 1 alpha chain;PDBTitle: the crystal structure of the narghi mutant nari-k86a
20 d1dmra2
97.7
21
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
21 c1tmoA_
not modelled
97.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: trimethylamine n-oxide reductase;PDBTitle: trimethylamine n-oxide reductase from shewanella massilia
22 c1eu1A_
not modelled
97.4
26
PDB header: oxidoreductaseChain: A: PDB Molecule: dimethyl sulfoxide reductase;PDBTitle: the crystal structure of rhodobacter sphaeroides dimethylsulfoxide2 reductase reveals two distinct molybdenum coordination environments.
23 d1tmoa2
not modelled
97.3
17
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
24 d1eu1a2
not modelled
97.2
26
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
25 c1vlfQ_
not modelled
97.1
28
PDB header: oxidoreductaseChain: Q: PDB Molecule: pyrogallol hydroxytransferase large subunit;PDBTitle: crystal structure of pyrogallol-phloroglucinol transhydroxylase from2 pelobacter acidigallici complexed with inhibitor 1,2,4,5-3 tetrahydroxy-benzene
26 c6cz7C_
not modelled
97.1
20
PDB header: oxidoreductaseChain: C: PDB Molecule: arra;PDBTitle: the arsenate respiratory reductase (arr) complex from shewanella sp.2 ana-3
27 c1h5nC_
not modelled
96.7
25
PDB header: oxidoreductaseChain: C: PDB Molecule: dmso reductase;PDBTitle: dmso reductase modified by the presence of dms and air
28 c2e7zA_
not modelled
96.3
25
PDB header: lyaseChain: A: PDB Molecule: acetylene hydratase ahy;PDBTitle: acetylene hydratase from pelobacter acetylenicus
29 d1vlfm2
not modelled
95.9
17
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
30 c5t5mB_
not modelled
88.0
24
PDB header: oxidoreductaseChain: B: PDB Molecule: tungsten formylmethanofuran dehydrogenase subunit fwdb;PDBTitle: tungsten-containing formylmethanofuran dehydrogenase from2 methanothermobacter wolfeii, trigonal form at 2.5 a.
31 d1ja1a1
not modelled
58.5
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: NADPH-cytochrome p450 reductase FAD-binding domain-like
32 c2ev2B_
not modelled
48.6
13
PDB header: lyaseChain: B: PDB Molecule: hypothetical protein rv1264/mt1302;PDBTitle: structure of rv1264n, the regulatory domain of the mycobacterial2 adenylyl cylcase rv1264, at ph 8.5
33 d1jb9a1
not modelled
43.8
23
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
34 d1ddga1
not modelled
42.6
31
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: NADPH-cytochrome p450 reductase FAD-binding domain-like
35 c1l0oC_
not modelled
39.2
26
PDB header: protein bindingChain: C: PDB Molecule: sigma factor;PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
36 d1l0oc_
not modelled
39.2
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
37 c2kvtA_
not modelled
34.7
54
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yaia;PDBTitle: solution nmr structure of yaia from escherichia eoli. northeast2 structural genomics target er244
38 d1f20a1
not modelled
32.4
33
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: NADPH-cytochrome p450 reductase FAD-binding domain-like
39 d1ku2a1
not modelled
26.8
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain
40 d1r8da_
not modelled
26.1
15
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA-binding N-terminal domain of transcription activators
41 d2fug32
not modelled
23.0
15
Fold: Formate dehydrogenase/DMSO reductase, domains 1-3Superfamily: Formate dehydrogenase/DMSO reductase, domains 1-3Family: Formate dehydrogenase/DMSO reductase, domains 1-3
42 c6j7aB_
not modelled
21.2
20
PDB header: oxidoreductaseChain: B: PDB Molecule: heme oxygenase 1,nadph--cytochrome p450 reductase;PDBTitle: fusion protein of heme oxygenase-1 and nadph cytochrome p450 reductase2 (17aa)
43 c6nu9A_
not modelled
20.7
32
PDB header: viral proteinChain: A: PDB Molecule: zinc-binding non-structural protein;PDBTitle: crystal structure of a zinc-binding non-structural protein from the2 hepatitis e virus
44 c2bpoA_
not modelled
19.7
23
PDB header: reductaseChain: A: PDB Molecule: nadph-cytochrom p450 reductase;PDBTitle: crystal structure of the yeast cpr triple mutant: d74g, y75f, k78a.
45 c3qftA_
not modelled
18.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: nadph--cytochrome p450 reductase;PDBTitle: crystal structure of nadph-cytochrome p450 reductase (fad/nadph domain2 and r457h mutant)
46 c2qtzA_
not modelled
17.7
20
PDB header: oxidoreductaseChain: A: PDB Molecule: methionine synthase reductase;PDBTitle: crystal structure of the nadp+-bound fad-containing fnr-like module of2 human methionine synthase reductase
47 c3hh0C_
not modelled
17.4
0
PDB header: transcription regulatorChain: C: PDB Molecule: transcriptional regulator, merr family;PDBTitle: crystal structure of a transcriptional regulator, merr family from2 bacillus cereus
48 d2bw3a1
not modelled
16.3
33
Fold: Hermes dimerisation domainSuperfamily: Hermes dimerisation domainFamily: Hermes dimerisation domain
49 c6amaO_
not modelled
15.1
21
PDB header: dna binding protein/dnaChain: O: PDB Molecule: putative dna-binding protein;PDBTitle: structure of s. coelicolor/s. venezuelae bldc-smea-ssfa complex to2 3.09 angstrom
50 c3mqiA_
not modelled
14.1
26
PDB header: transcription activatorChain: A: PDB Molecule: transcription factor coe1;PDBTitle: human early b-cell factor 1 (ebf1) ipt/tig domain
51 c1y6uA_
not modelled
14.0
13
PDB header: dna binding proteinChain: A: PDB Molecule: excisionase from transposon tn916;PDBTitle: the structure of the excisionase (xis) protein from2 conjugative transposon tn916 provides insights into the3 regulation of heterobivalent tyrosine recombinases
52 c1f20A_
not modelled
13.6
37
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase fad/nadp+2 domain at 1.9a resolution.
53 c6efvA_
not modelled
13.0
31
PDB header: flavoproteinChain: A: PDB Molecule: sulfite reductase [nadph] flavoprotein alpha-component;PDBTitle: the nadph-dependent sulfite reductase flavoprotein adopts an extended2 conformation that is unique to this diflavin reductase
54 c4ediC_
not modelled
12.0
18
PDB header: transport proteinChain: C: PDB Molecule: ethanolamine utilization protein;PDBTitle: disulfide bonded eutl from clostridium perfringens
55 d1qfza1
not modelled
11.8
42
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like
56 c5o5jE_
not modelled
11.8
22
PDB header: ribosomeChain: E: PDB Molecule: 30s ribosomal protein s5;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
57 c1ifpA_
not modelled
11.1
39
PDB header: virusChain: A: PDB Molecule: major coat protein assembly;PDBTitle: inovirus (filamentous bacteriophage) strain pf3 major coat2 protein assembly
58 c1tllA_
not modelled
10.7
37
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase, brain;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase2 reductase module at 2.3 a resolution.
59 c3t0yA_
not modelled
10.1
12
PDB header: transcription regulator/protein bindingChain: A: PDB Molecule: response regulator;PDBTitle: structure of the phyr anti-anti-sigma domain bound to the anti-sigma2 factor, nepr
60 c3ezfA_
not modelled
9.7
13
PDB header: biosynthetic proteinChain: A: PDB Molecule: para;PDBTitle: partition protein
61 c1ddiA_
not modelled
9.6
27
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase [nadph] flavoprotein alpha-component;PDBTitle: crystal structure of sir-fp60
62 c2jmlA_
not modelled
9.3
15
PDB header: transcriptionChain: A: PDB Molecule: dna binding domain/transcriptional regulator;PDBTitle: solution structure of the n-terminal domain of cara repressor
63 c4l4qA_
not modelled
8.9
67
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: methionine adenosyltransferase
64 c2dqbB_
not modelled
8.6
26
PDB header: hydrolase, dna binding proteinChain: B: PDB Molecule: deoxyguanosinetriphosphate triphosphohydrolase, putative;PDBTitle: crystal structure of dntp triphosphohydrolase from thermus2 thermophilus hb8, which is homologous to dgtp triphosphohydrolase
65 c3dupB_
not modelled
8.4
17
PDB header: hydrolaseChain: B: PDB Molecule: mutt/nudix family protein;PDBTitle: crystal structure of mutt/nudix family hydrolase from rhodospirillum2 rubrum atcc 11170
66 c2fugC_
not modelled
8.1
16
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh-quinone oxidoreductase chain 3;PDBTitle: crystal structure of the hydrophilic domain of respiratory complex i2 from thermus thermophilus
67 c4dqkA_
not modelled
8.1
25
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional p-450/nadph-p450 reductase;PDBTitle: crystal structure of the fad binding domain of cytochrome p450 bm3
68 c3io0A_
not modelled
8.0
36
PDB header: structural proteinChain: A: PDB Molecule: etub protein;PDBTitle: crystal structure of etub from clostridium kluyveri
69 d2p7vb1
not modelled
7.9
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain
70 c1dpuA_
not modelled
7.3
15
PDB header: dna binding proteinChain: A: PDB Molecule: replication protein a (rpa32) c-terminal domain;PDBTitle: solution structure of the c-terminal domain of human rpa322 complexed with ung2(73-88)
71 d1dpua_
not modelled
7.3
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: C-terminal domain of RPA32
72 c4cc9B_
not modelled
7.3
20
PDB header: protein bindingChain: B: PDB Molecule: protein vpx;PDBTitle: crystal structure of human samhd1 (amino acid residues 582-626) bound2 to vpx isolated from sooty mangabey and human dcaf1 (amino acid3 residues 1058-1396)
73 c4hpvA_
not modelled
7.1
67
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: crystal structure of s-adenosylmethionine synthetase from sulfolobus2 solfataricus
74 c4fayC_
not modelled
7.0
24
PDB header: glycerol-binding proteinChain: C: PDB Molecule: microcompartments protein;PDBTitle: crystal structure of a trimeric bacterial microcompartment shell2 protein pdub with glycerol metabolites
75 c1j9zB_
not modelled
7.0
20
PDB header: oxidoreductaseChain: B: PDB Molecule: nadph-cytochrome p450 reductase;PDBTitle: cypor-w677g
76 c4r4eA_
not modelled
6.8
17
PDB header: transcription regulator/dnaChain: A: PDB Molecule: hth-type transcriptional regulator glnr;PDBTitle: structure of glnr-dna complex
77 c2lfwA_
not modelled
6.7
12
PDB header: signaling proteinChain: A: PDB Molecule: phyr sigma-like domain;PDBTitle: nmr structure of the phyrsl-nepr complex from sphingomonas sp. fr1
78 c2vz4A_
not modelled
6.6
11
PDB header: transcriptionChain: A: PDB Molecule: hth-type transcriptional activator tipa;PDBTitle: the n-terminal domain of merr-like protein tipal bound to promoter dna
79 c5af3A_
not modelled
6.5
6
PDB header: dna bindingChain: A: PDB Molecule: vapbc49;PDBTitle: x-ray crystal structure of rv2018 from mycobacterium tuberculosis
80 c5c8eC_
not modelled
6.4
25
PDB header: transcription regulator/dnaChain: C: PDB Molecule: light-dependent transcriptional regulator carh;PDBTitle: crystal structure of thermus thermophilus carh bound to2 adenosylcobalamin and a 26-bp dna segment
81 c5u4nA_
not modelled
6.1
25
PDB header: lyaseChain: A: PDB Molecule: fructose-1;PDBTitle: crystal structure of a fructose-bisphosphate aldolase from neisseria2 gonorrhoeae
82 c3u27D_
not modelled
6.1
19
PDB header: structural proteinChain: D: PDB Molecule: microcompartments protein;PDBTitle: crystal structure of ethanolamine utilization protein eutl from2 leptotrichia buccalis c-1013-b
83 c5z9aB_
not modelled
6.0
31
PDB header: lyaseChain: B: PDB Molecule: chorismate synthase;PDBTitle: crystal structure of chorismate synthase from pseudomonas aeruginosa
84 c3iwfA_
not modelled
5.7
6
PDB header: transcription regulatorChain: A: PDB Molecule: transcription regulator rpir family;PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
85 c5i44E_
not modelled
5.4
17
PDB header: dna binding protein/dnaChain: E: PDB Molecule: chromosome-anchoring protein raca;PDBTitle: structure of raca-dna complex; p21 form