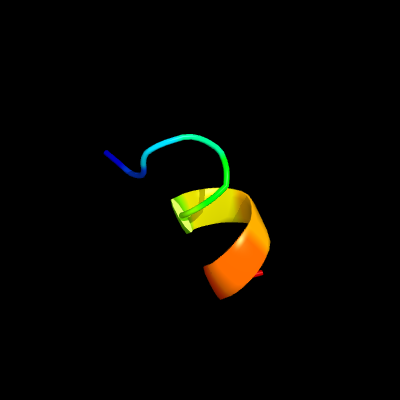1 c2f5zK_
41.6
73
PDB header: oxidoreductase/protein bindingChain: K: PDB Molecule: pyruvate dehydrogenase protein x component;PDBTitle: crystal structure of human dihydrolipoamide dehydrogenase2 (e3) complexed to the e3-binding domain of human e3-3 binding protein
2 c1zy8M_
40.6
73
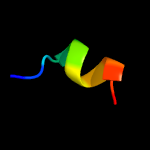
PDB header: oxidoreductaseChain: M: PDB Molecule: pyruvate dehydrogenase protein x component,PDBTitle: the crystal structure of dihydrolipoamide dehydrogenase and2 dihydrolipoamide dehydrogenase-binding protein (didomain)3 subcomplex of human pyruvate dehydrogenase complex.
3 c2f60K_
31.3
64
PDB header: protein bindingChain: K: PDB Molecule: pyruvate dehydrogenase protein x component;PDBTitle: crystal structure of the dihydrolipoamide dehydrogenase (e3)-binding2 domain of human e3-binding protein
4 c5h0jA_
25.6
47
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of wt pedobacter heparinus smug2
5 c3kwvF_
23.4
38
PDB header: toxin/protein transportChain: F: PDB Molecule: lethal factor;PDBTitle: structural basis for the unfolding of anthrax lethal factor by2 protective antigen oligomers
6 c3ah5E_
21.6
17
PDB header: transferaseChain: E: PDB Molecule: thymidylate synthase thyx;PDBTitle: crystal structure of flavin dependent thymidylate synthase thyx from2 helicobacter pylori complexed with fad and dump
7 d1j7na1
21.4
38
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Anthrax toxin lethal factor, N- and C-terminal domains
8 c4pugA_
19.6
20
PDB header: dna binding proteinChain: A: PDB Molecule: bola like protein;PDBTitle: bola1 from arabidopsis thaliana
9 c5x42B_
17.7
54
PDB header: protein transportChain: B: PDB Molecule: icmo (dotl);PDBTitle: structure of dotl(590-659)-dotn derived from legionella pneumophila
10 d1oe4a_
16.2
31
Fold: Uracil-DNA glycosylase-likeSuperfamily: Uracil-DNA glycosylase-likeFamily: Single-strand selective monofunctional uracil-DNA glycosylase SMUG1
11 c3o2eA_
16.0
29
PDB header: unknown functionChain: A: PDB Molecule: bola-like protein;PDBTitle: crystal structure of a bol-like protein from babesia bovis
12 c5nfmA_
14.7
24
PDB header: ligaseChain: A: PDB Molecule: yrba;PDBTitle: crystal structure of yrba from sinorhizobium meliloti in complex with2 copper.
13 c5h93C_
14.3
53
PDB header: hydrolaseChain: C: PDB Molecule: geobacter metallireducens smug1;PDBTitle: crystal structure of geobacter metallireducens smug1
14 c2hacA_
14.1
63
PDB header: membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd3 zeta chain;PDBTitle: structure of zeta-zeta transmembrane dimer
15 c2hacB_
14.1
63
PDB header: membrane proteinChain: B: PDB Molecule: t-cell surface glycoprotein cd3 zeta chain;PDBTitle: structure of zeta-zeta transmembrane dimer
16 c5w6yB_
14.0
64
PDB header: biosynthetic protein,isomeraseChain: B: PDB Molecule: chorismate mutase;PDBTitle: physcomitrella patens chorismate mutase
17 c2q2kB_
13.9
48
PDB header: dna binding protein/dnaChain: B: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
18 c2q2kA_
12.8
48
PDB header: dna binding protein/dnaChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
19 c6h3pB_
12.1
55
PDB header: plant proteinChain: B: PDB Molecule: chorismate mutase;PDBTitle: crystal structure of the cytoplasmic chorismate mutase from zea mays
20 d5csma_
12.1
27
Fold: Chorismate mutase IISuperfamily: Chorismate mutase IIFamily: Allosteric chorismate mutase
21 c2af6G_
not modelled
12.1
33
PDB header: transferaseChain: G: PDB Molecule: thymidylate synthase thyx;PDBTitle: crystal structure of mycobacterium tuberculosis flavin dependent2 thymidylate synthase (mtb thyx) in the presence of co-factor fad and3 substrate analog 5-bromo-2'-deoxyuridine-5'-monophosphate (brdump)
22 c4ppuA_
not modelled
12.0
55
PDB header: isomeraseChain: A: PDB Molecule: chorismate mutase 1, chloroplastic;PDBTitle: crystal structure of atcm1 with tyrosine bound in allosteric site
23 c5w8cA_
not modelled
11.4
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: autoinducer synthase;PDBTitle: the structure of a coa-dependent acyl-homoserine lactone synthase,2 bjai, with mta and isovaleryl-coa
24 d1pv8a_
not modelled
11.0
22
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
25 c5uebA_
not modelled
10.3
22
PDB header: unknown functionChain: A: PDB Molecule: negoa.19184.a;PDBTitle: novel crystal structure of a hypothetical protein from neisseria2 gonorrhoeae
26 c2dhmA_
not modelled
9.1
17
PDB header: protein bindingChain: A: PDB Molecule: protein bola;PDBTitle: solution structure of the bola protein from escherichia coli
27 c3tr3A_
not modelled
9.0
31
PDB header: unknown functionChain: A: PDB Molecule: bola;PDBTitle: structure of a bola protein homologue from coxiella burnetii
28 c1v60A_
not modelled
8.9
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: riken cdna 1810037g04;PDBTitle: solution structure of bola1 protein from mus musculus
29 c3fnnA_
not modelled
8.7
27
PDB header: transferaseChain: A: PDB Molecule: thymidylate synthase thyx;PDBTitle: biochemical and structural analysis of an atypical thyx:2 corynebacterium glutamicum nchu 87078 depends on thya for3 thymidine biosynthesis
30 c3obkH_
not modelled
8.3
23
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
31 d2c1ha1
not modelled
7.9
27
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
32 c4k59A_
not modelled
7.8
43
PDB header: rna binding proteinChain: A: PDB Molecule: rna binding protein rsmf;PDBTitle: crystal structure of pseudomonas aeruginosa rsmf
33 c1xs3A_
not modelled
7.5
34
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein xc975;PDBTitle: solution structure analysis of the xc975 protein
34 c1xfzA_
not modelled
7.2
31
PDB header: lyase/metal binding proteinChain: A: PDB Molecule: calmodulin-sensitive adenylate cyclase;PDBTitle: crystal structure of anthrax edema factor (ef) in complex with2 calmodulin in the presence of 1 millimolar exogenously added calcium3 chloride
35 c1xfuE_
not modelled
7.2
31
PDB header: lyase/metal binding proteinChain: E: PDB Molecule: calmodulin-sensitive adenylate cyclase;PDBTitle: crystal structure of anthrax edema factor (ef) truncation mutant, ef-2 delta 64 in complex with calmodulin
36 d1h7na_
not modelled
7.2
27
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
37 c4puiA_
not modelled
6.7
34
PDB header: protein bindingChain: A: PDB Molecule: sufe-like protein, chloroplastic;PDBTitle: bola domain of sufe1 from arabidopsis thaliana
38 c6fpgG_
not modelled
6.4
29
PDB header: cell invasionChain: G: PDB Molecule: chromosome 16, whole genome shotgun sequence;PDBTitle: structure of the ustilago maydis chorismate mutase 1 in complex with a2 zea mays kiwellin
39 d2csua1
not modelled
6.3
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
40 c2mm9A_
not modelled
6.2
43
PDB header: transcriptionChain: A: PDB Molecule: bola2;PDBTitle: solution structure of reduced bola2 from arabidopsis thaliana
41 c2nclA_
not modelled
5.7
14
PDB header: protein bindingChain: A: PDB Molecule: bola-like protein 3;PDBTitle: solution structure of bola3 from homo sapiens
42 d1gzga_
not modelled
5.7
28
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
43 c3gmvX_
not modelled
5.6
40
PDB header: protein bindingChain: X: PDB Molecule: beta-lactamase inhibitory protein blip-i;PDBTitle: crystal structure of beta-lactamse inhibitory protein-i (blip-i) in2 apo form
44 d1coka_
not modelled
5.6
11
Fold: SAM domain-likeSuperfamily: SAM/Pointed domainFamily: SAM (sterile alpha motif) domain
45 c3gmxB_
not modelled
5.5
53
PDB header: protein bindingChain: B: PDB Molecule: blp;PDBTitle: crystal structure of beta-lactamse inhibitory protein-like protein2 (blp) at 1.05 angstrom resolution
46 c1pwqA_
not modelled
5.4
38
PDB header: hydrolaseChain: A: PDB Molecule: lethal factor;PDBTitle: crystal structure of anthrax lethal factor complexed with2 thioacetyl-tyr-pro-met-amide, a metal-chelating peptidyl3 small molecule inhibitor
47 d2py5a2
not modelled
5.3
41
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: DNA polymerase I
48 d1l6sa_
not modelled
5.2
25
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
49 c5lzlH_
not modelled
5.1
32
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase
50 c3pmkR_
not modelled
5.1
53
PDB header: viral proteinChain: R: PDB Molecule: phosphoprotein;PDBTitle: crystal structure of the vesicular stomatitis virus rna free2 nucleoprotein/phosphoprotein complex
51 c3pmkP_
not modelled
5.1
53
PDB header: viral proteinChain: P: PDB Molecule: phosphoprotein;PDBTitle: crystal structure of the vesicular stomatitis virus rna free2 nucleoprotein/phosphoprotein complex
52 c5dcaA_
not modelled
5.0
23
PDB header: hydrolaseChain: A: PDB Molecule: pre-mrna-splicing helicase brr2;PDBTitle: crystal structure of yeast full length brr2 in complex with prp8 jab12 domain
53 c5oooA_
not modelled
5.0
34
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein ns-s;PDBTitle: structure of the rift valley fever virus nss protein core domain
54 c3pmkN_
not modelled
5.0
53
PDB header: viral proteinChain: N: PDB Molecule: phosphoprotein;PDBTitle: crystal structure of the vesicular stomatitis virus rna free2 nucleoprotein/phosphoprotein complex