1 c6h77B_
100.0
22
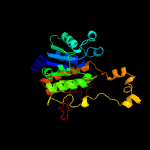
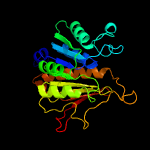
PDB header: signaling proteinChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme 5;PDBTitle: e1 enzyme for ubiquitin like protein activation in complex with ubl
2 d1yovb1
100.0
23
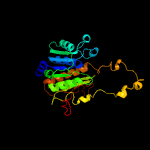
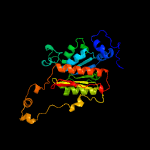
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)
3 c1zfnA_
100.0
25
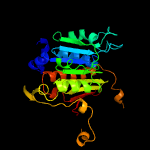
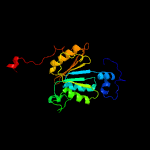
PDB header: transferaseChain: A: PDB Molecule: adenylyltransferase thif;PDBTitle: structural analysis of escherichia coli thif
4 c3h9gA_
100.0
19
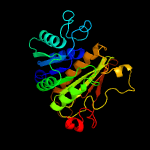
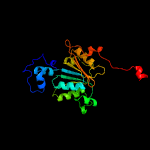
PDB header: transferase/antibioticChain: A: PDB Molecule: mccb protein;PDBTitle: crystal structure of e. coli mccb + mcca-n7isoasn
5 d1jw9b_
100.0
23
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Molybdenum cofactor biosynthesis protein MoeB
6 c3gznB_
100.0
23
PDB header: protein binding/ligaseChain: B: PDB Molecule: nedd8-activating enzyme e1 catalytic subunit;PDBTitle: structure of nedd8-activating enzyme in complex with nedd8 and mln4924
7 c5ff5A_
100.0
19
PDB header: transferaseChain: A: PDB Molecule: paaa;PDBTitle: crystal structure of semet paaa
8 c4d7aA_
100.0
23
PDB header: ligaseChain: A: PDB Molecule: trna threonylcarbamoyladenosine dehydratase;PDBTitle: crystal structure of e. coli trna n6-threonylcarbamoyladenosine2 dehydratase, tcda, in complex with amp at 1.801 angstroem3 resolution
9 c2nvuB_
100.0
22
PDB header: protein turnover, ligaseChain: B: PDB Molecule: maltose binding protein/nedd8-activating enzymePDBTitle: structure of appbp1-uba3~nedd8-nedd8-mgatp-ubc12(c111a), a2 trapped ubiquitin-like protein activation complex
10 c3kydB_
100.0
22
PDB header: ligaseChain: B: PDB Molecule: sumo-activating enzyme subunit 2;PDBTitle: human sumo e1~sumo1-amp tetrahedral intermediate mimic
11 c3kycB_
100.0
23
PDB header: ligaseChain: B: PDB Molecule: sumo-activating enzyme subunit 2;PDBTitle: human sumo e1 complex with a sumo1-amp mimic
12 c1y8qD_
100.0
24
PDB header: ligaseChain: D: PDB Molecule: ubiquitin-like 2 activating enzyme e1b;PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
13 c1y8qA_
100.0
18
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-like 1 activating enzyme e1a;PDBTitle: sumo e1 activating enzyme sae1-sae2-mg-atp complex
14 c3vh1A_
100.0
18
PDB header: metal binding proteinChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of saccharomyces cerevisiae atg7 (1-595)
15 c3vh3A_
100.0
17
PDB header: metal binding protein/protein transportChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme atg7;PDBTitle: crystal structure of atg7ctd-atg8 complex
16 c3cmmA_
100.0
28
PDB header: ligase/protein bindingChain: A: PDB Molecule: ubiquitin-activating enzyme e1 1;PDBTitle: crystal structure of the uba1-ubiquitin complex
17 c6dc6A_
100.0
20
PDB header: signaling protein/ligaseChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme 1;PDBTitle: crystal structure of human ubiquitin activating enzyme e1 (uba1) in2 complex with ubiquitin
18 c4ii3A_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-activating enzyme e1 1;PDBTitle: crystal structure of s. pombe ubiquitin activating enzyme 1 (uba1) in2 complex with ubiquitin and atp/mg
19 d1yova1
100.0
21
Fold: Activating enzymes of the ubiquitin-like proteinsSuperfamily: Activating enzymes of the ubiquitin-like proteinsFamily: Ubiquitin activating enzymes (UBA)
20 c4p22A_
100.0
22
PDB header: ligaseChain: A: PDB Molecule: ubiquitin-like modifier-activating enzyme 1;PDBTitle: crystal structure of n-terminal fragments of e1
21 c3wv9D_
not modelled
100.0
18
PDB header: transferaseChain: D: PDB Molecule: hmd co-occurring protein hcge;PDBTitle: guanylylpyridinol (gp)- and atp-bound hcge from methanothermobacter2 marburgensis
22 c3gucB_
not modelled
100.0
19
PDB header: transferaseChain: B: PDB Molecule: ubiquitin-like modifier-activating enzyme 5;PDBTitle: human ubiquitin-activating enzyme 5 in complex with amppnp
23 c4rl6A_
not modelled
98.7
13
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q04l03_strp2 protein from streptococcus2 pneumoniae. northeast structural genomics consortium target spr105
24 c1e5lA_
not modelled
98.7
17
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine reductase;PDBTitle: apo saccharopine reductase from magnaporthe grisea
25 c2axqA_
not modelled
98.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: apo histidine-tagged saccharopine dehydrogenase (l-glu2 forming) from saccharomyces cerevisiae
26 c5l78A_
not modelled
98.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: alpha-aminoadipic semialdehyde synthase, mitochondrial;PDBTitle: crystal structure of human aminoadipate semialdehyde synthase,2 saccharopine dehydrogenase domain (in nad+ bound form)
27 c4bs9A_
not modelled
98.5
18
PDB header: isomeraseChain: A: PDB Molecule: trud;PDBTitle: structure of the heterocyclase trud
28 c3ic5A_
not modelled
98.3
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative saccharopine dehydrogenase;PDBTitle: n-terminal domain of putative saccharopine dehydrogenase from ruegeria2 pomeroyi.
29 d1vi2a1
not modelled
98.3
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
30 c4inaA_
not modelled
98.2
17
PDB header: oxidoreductaseChain: A: PDB Molecule: saccharopine dehydrogenase;PDBTitle: crystal structure of the q7mss8_wolsu protein from wolinella2 succinogenes. northeast structural genomics consortium target wsr35
31 c3tozA_
not modelled
98.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: 2.2 angstrom crystal structure of shikimate 5-dehydrogenase from2 listeria monocytogenes in complex with nad.
32 d1pjqa1
not modelled
98.1
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Siroheme synthase N-terminal domain-like
33 c2z2vA_
not modelled
98.1
16
PDB header: oxidoreductaseChain: A: PDB Molecule: hypothetical protein ph1688;PDBTitle: crystal structure of l-lysine dehydrogenase from2 hyperthermophilic archaeon pyrococcus horikoshii
34 c1vi2B_
not modelled
98.1
18
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase 2;PDBTitle: crystal structure of shikimate-5-dehydrogenase with nad
35 c2nloA_
not modelled
98.0
22
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of the quinate dehydrogenase from corynebacterium2 glutamicum
36 c4plpB_
not modelled
98.0
17
PDB header: transferaseChain: B: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase (hss) from2 blastochloris viridis in complex with nad
37 c1gpjA_
not modelled
97.9
20
PDB header: reductaseChain: A: PDB Molecule: glutamyl-trna reductase;PDBTitle: glutamyl-trna reductase from methanopyrus kandleri
38 d1e5qa1
not modelled
97.9
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain
39 d2pgda2
not modelled
97.9
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
40 c3pgjB_
not modelled
97.9
19
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate dehydrogenase;PDBTitle: 2.49 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae o1 biovar eltor str. n169613 in complex with shikimate
41 c2eggA_
not modelled
97.9
22
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate 5-dehydrogenase;PDBTitle: crystal structure of shikimate 5-dehydrogenase (aroe) from geobacillus2 kaustophilus
42 d1pzga1
not modelled
97.9
19
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
43 c3tumA_
not modelled
97.9
27
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase family protein;PDBTitle: 2.15 angstrom resolution crystal structure of a shikimate2 dehydrogenase family protein from pseudomonas putida kt2440 in3 complex with nad+
44 c4n7rB_
not modelled
97.8
22
PDB header: oxidoreductase/protein bindingChain: B: PDB Molecule: glutamyl-trna reductase 1, chloroplastic;PDBTitle: crystal structure of arabidopsis glutamyl-trna reductase in complex2 with its binding protein
45 d1uxja1
not modelled
97.8
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
46 d2nu7a1
not modelled
97.7
13
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
47 c3u62A_
not modelled
97.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from thermotoga maritima
48 c3o8qB_
not modelled
97.7
23
PDB header: oxidoreductaseChain: B: PDB Molecule: shikimate 5-dehydrogenase i alpha;PDBTitle: 1.45 angstrom resolution crystal structure of shikimate 5-2 dehydrogenase (aroe) from vibrio cholerae
49 d1oi7a1
not modelled
97.7
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
50 c2hjrK_
not modelled
97.7
18
PDB header: oxidoreductaseChain: K: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of cryptosporidium parvum malate2 dehydrogenase
51 c6iauB_
not modelled
97.6
25
PDB header: oxidoreductaseChain: B: PDB Molecule: amine dehydrogenase;PDBTitle: amine dehydrogenase from cystobacter fuscus in complex with nadp+ and2 cyclohexylamine
52 c4bgvB_
not modelled
97.6
24
PDB header: oxidoreductaseChain: B: PDB Molecule: malate dehydrogenase;PDBTitle: 1.8 a resolution structure of the malate dehydrogenase from2 picrophilus torridus in its apo form
53 c1npyA_
not modelled
97.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical shikimate 5-dehydrogenase-like protein hi0607;PDBTitle: structure of shikimate 5-dehydrogenase-like protein hi0607
54 c1pjtB_
not modelled
97.6
19
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
55 c3donA_
not modelled
97.6
22
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase from staphylococcus2 epidermidis
56 c1pgqA_
not modelled
97.6
18
PDB header: oxidoreductase (choh(d)-nadp+(a))Chain: A: PDB Molecule: 6-phosphogluconate dehydrogenase;PDBTitle: crystallographic study of coenzyme, coenzyme analogue and substrate2 binding in 6-phosphogluconate dehydrogenase: implications for nadp3 specificity and the enzyme mechanism
57 d9ldta1
not modelled
97.6
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
58 d1gpja2
not modelled
97.6
23
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
59 d1pgja2
not modelled
97.6
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
60 c2ph5A_
not modelled
97.5
19
PDB header: transferaseChain: A: PDB Molecule: homospermidine synthase;PDBTitle: crystal structure of the homospermidine synthase hss from legionella2 pneumophila in complex with nad, northeast structural genomics target3 lgr54
61 c6fqzB_
not modelled
97.5
14
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: plasmodium falciparum 6-phosphogluconate dehydrogenase in its apo2 form, in complex with its cofactor nadp+ and in complex with its3 substrate 6-phosphogluconate
62 c3fwnB_
not modelled
97.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: dimeric 6-phosphogluconate dehydrogenase complexed with 6-2 phosphogluconate and 2'-monophosphoadenosine-5'-diphosphate
63 c2iz1C_
not modelled
97.5
15
PDB header: oxidoreductaseChain: C: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating;PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
64 c4tskA_
not modelled
97.5
25
PDB header: oxidoreductase,isomeraseChain: A: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: ketol-acid reductoisomerase from alicyclobacillus acidocaldarius
65 d1pjca1
not modelled
97.5
18
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Formate/glycerate dehydrogenases, NAD-domain
66 d1npya1
not modelled
97.5
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
67 c4xijA_
not modelled
97.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate 5-dehydrogenase;PDBTitle: crystal structure of a shikimate 5-dehydrogenase from mycobacterium2 fortuitum determined by iodide sad phasing
68 c2fnzA_
not modelled
97.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of the lactate dehydrogenase from cryptosporidium2 parvum complexed with cofactor (b-nicotinamide adenine dinucleotide)3 and inhibitor (oxamic acid)
69 c4ypoB_
not modelled
97.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: crystal structure of mycobacterium tuberculosis ketol-acid2 reductoisomerase in complex with mg2+
70 c1pgjA_
not modelled
97.5
18
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase;PDBTitle: x-ray structure of 6-phosphogluconate dehydrogenase from the protozoan2 parasite t. brucei
71 c4e21B_
not modelled
97.5
17
PDB header: oxidoreductaseChain: B: PDB Molecule: 6-phosphogluconate dehydrogenase (decarboxylating);PDBTitle: the crystal structure of 6-phosphogluconate dehydrogenase from2 geobacter metallireducens
72 c3pvzD_
not modelled
97.5
14
PDB header: lyaseChain: D: PDB Molecule: udp-n-acetylglucosamine 4,6-dehydratase;PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri
73 d1i0za1
not modelled
97.5
12
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
74 c5swvC_
not modelled
97.5
12
PDB header: lyaseChain: C: PDB Molecule: pentafunctional arom polypeptide;PDBTitle: dehydroquinate dehydratase and shikimate dehydrogenase from s. pombe2 arom
75 c4ywjB_
not modelled
97.5
21
PDB header: oxidoreductaseChain: B: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of 4-hydroxy-tetrahydrodipicolinate reductase (htpa2 reductase) from pseudomonas aeruginosa
76 c3pwzA_
not modelled
97.5
29
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase 3;PDBTitle: crystal structure of an ael1 enzyme from pseudomonas putida
77 d1u8xx1
not modelled
97.5
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
78 c4kqxB_
not modelled
97.5
16
PDB header: oxidoreductase/oxidoreductase inhibitorChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: mutant slackia exigua kari ddv in complex with nad and an inhibitor
79 d1np3a2
not modelled
97.5
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain
80 c1pzfD_
not modelled
97.5
25
PDB header: oxidoreductaseChain: D: PDB Molecule: lactate dehydrogenase;PDBTitle: t.gondii ldh1 ternary complex with apad+ and oxalate
81 c4xdzB_
not modelled
97.5
18
PDB header: oxidoreductaseChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: holo structure of ketol-acid reductoisomerase from ignisphaera2 aggregans
82 c5yeqB_
not modelled
97.5
15
PDB header: isomeraseChain: B: PDB Molecule: ketol-acid reductoisomerase (nadp(+));PDBTitle: the structure of sac-kari protein
83 c3nzoB_
not modelled
97.5
14
PDB header: lyaseChain: B: PDB Molecule: udp-n-acetylglucosamine 4,6-dehydratase;PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri.
84 c1bg6A_
not modelled
97.4
17
PDB header: oxidoreductaseChain: A: PDB Molecule: n-(1-d-carboxylethyl)-l-norvaline dehydrogenase;PDBTitle: crystal structure of the n-(1-d-carboxylethyl)-l-norvaline2 dehydrogenase from arthrobacter sp. strain 1c
85 c3k6jA_
not modelled
97.4
14
PDB header: oxidoreductaseChain: A: PDB Molecule: protein f01g10.3, confirmed by transcript evidence;PDBTitle: crystal structure of the dehydrogenase part of multifuctional enzyme 12 from c.elegans
86 c4omuA_
not modelled
97.4
24
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase;PDBTitle: crystal structure of shikimate dehydrogenase (aroe) from pseudomonas2 putida
87 d1euca1
not modelled
97.4
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: CoA-binding domain
88 c1ldbA_
not modelled
97.4
25
PDB header: oxidoreductase(choh(d)-nad(a))Chain: A: PDB Molecule: apo-l-lactate dehydrogenase;PDBTitle: structure determination and refinement of bacillus2 stearothermophilus lactate dehydrogenase
89 c5nfrI_
not modelled
97.4
13
PDB header: oxidoreductaseChain: I: PDB Molecule: malate dehydrogenase;PDBTitle: crystal structure of malate dehydrogenase from plasmodium falciparum2 (pfmdh)
90 c5dzsA_
not modelled
97.4
16
PDB header: oxidoreductaseChain: A: PDB Molecule: shikimate dehydrogenase (nadp(+));PDBTitle: 1.5 angstrom crystal structure of shikimate dehydrogenase 1 from2 peptoclostridium difficile.
91 c2p4qA_
not modelled
97.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase, decarboxylating 1;PDBTitle: crystal structure analysis of gnd1 in saccharomyces cerevisiae
92 c4ezbA_
not modelled
97.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein;PDBTitle: crystal structure of the conserved hypothetical protein from2 sinorhizobium meliloti 1021
93 c1np3B_
not modelled
97.4
23
PDB header: oxidoreductaseChain: B: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: crystal structure of class i acetohydroxy acid isomeroreductase from2 pseudomonas aeruginosa
94 c6iaqA_
not modelled
97.4
15
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase n-terminus domain-containingPDBTitle: structure of amine dehydrogenase from mycobacterium smegmatis
95 c1z7eC_
not modelled
97.4
21
PDB header: hydrolaseChain: C: PDB Molecule: protein arna;PDBTitle: crystal structure of full length arna
96 c4mp6A_
not modelled
97.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: putative ornithine cyclodeaminase;PDBTitle: staphyloferrin b precursor biosynthetic enzyme sbnb bound to citrate2 and nad+
97 c1u4sA_
not modelled
97.4
18
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase;PDBTitle: plasmodium falciparum lactate dehydrogenase complexed with 2,6-2 naphthalenedisulphonic acid
98 c5ugjC_
not modelled
97.3
20
PDB header: oxidoreductaseChain: C: PDB Molecule: 4-hydroxy-tetrahydrodipicolinate reductase;PDBTitle: crystal structure of htpa reductase from neisseria meningitidis
99 c4xcvA_
not modelled
97.3
20
PDB header: oxidoreductaseChain: A: PDB Molecule: nadp-dependent 2-hydroxyacid dehydrogenase;PDBTitle: probable 2-hydroxyacid dehydrogenase from rhizobium etli cfn 42 in2 complex with nadph
100 d1s6ya1
not modelled
97.3
17
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
101 c1drwA_
not modelled
97.3
21
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodipicolinate reductase;PDBTitle: escherichia coli dhpr/nhdh complex
102 c1m67A_
not modelled
97.3
11
PDB header: oxidoreductaseChain: A: PDB Molecule: glycerol-3-phosphate dehydrogenase;PDBTitle: crystal structure of leishmania mexicana gpdh complexed with inhibitor2 2-bromo-6-hydroxy-purine
103 d1i10a1
not modelled
97.3
9
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
104 d1nvta1
not modelled
97.3
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
105 c6ihdA_
not modelled
97.3
16
PDB header: oxidoreductaseChain: A: PDB Molecule: malate dehydrogenase;PDBTitle: crystal strcuture of malate dehydrogenase from metallosphaera sedula
106 c2dc1A_
not modelled
97.3
24
PDB header: oxidoreductaseChain: A: PDB Molecule: l-aspartate dehydrogenase;PDBTitle: crystal structure of l-aspartate dehydrogenase from2 hyperthermophilic archaeon archaeoglobus fulgidus
107 c1mldA_
not modelled
97.2
28
PDB header: oxidoreductase(nad(a)-choh(d))Chain: A: PDB Molecule: malate dehydrogenase;PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
108 c4plcA_
not modelled
97.2
18
PDB header: oxidoreductaseChain: A: PDB Molecule: lactate dehydrogenase;PDBTitle: crystal structure of ancestral apicomplexan lactate dehydrogenase with2 malate.
109 c3k96B_
not modelled
97.2
20
PDB header: oxidoreductaseChain: B: PDB Molecule: glycerol-3-phosphate dehydrogenase [nad(p)+];PDBTitle: 2.1 angstrom resolution crystal structure of glycerol-3-phosphate2 dehydrogenase (gpsa) from coxiella burnetii
110 d2ldxa1
not modelled
97.2
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
111 d2jfga1
not modelled
97.2
20
Fold: MurCD N-terminal domainSuperfamily: MurCD N-terminal domainFamily: MurCD N-terminal domain
112 d1obba1
not modelled
97.2
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
113 c2v65A_
not modelled
97.2
15
PDB header: oxidoreductaseChain: A: PDB Molecule: l-lactate dehydrogenase a chain;PDBTitle: apo ldh from the psychrophile c. gunnari
114 d5ldha1
not modelled
97.2
10
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: LDH N-terminal domain-like
115 c3db2C_
not modelled
97.2
15
PDB header: oxidoreductaseChain: C: PDB Molecule: putative nadph-dependent oxidoreductase;PDBTitle: crystal structure of a putative nadph-dependent oxidoreductase2 (dhaf_2064) from desulfitobacterium hafniense dcb-2 at 1.70 a3 resolution
116 c2qx7A_
not modelled
97.2
12
PDB header: plant proteinChain: A: PDB Molecule: eugenol synthase 1;PDBTitle: structure of eugenol synthase from ocimum basilicum
117 c5y8mA_
not modelled
97.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: probable 3-hydroxyisobutyrate dehydrogenase;PDBTitle: mycobacterium tuberculosis 3-hydroxyisobutyrate dehydrogenase2 (mthibadh) + nad + (r)-3-hydroxyisobutyrate (r-hiba)
118 c4n18A_
not modelled
97.1
26
PDB header: oxidoreductaseChain: A: PDB Molecule: d-isomer specific 2-hydroxyacid dehydrogenase familyPDBTitle: crystal structure of d-isomer specific 2-hydroxyacid dehydrogenase2 family protein from klebsiella pneumoniae 342
119 c3pqeD_
not modelled
97.1
28
PDB header: oxidoreductaseChain: D: PDB Molecule: l-lactate dehydrogenase;PDBTitle: crystal structure of l-lactate dehydrogenase from bacillus subtilis2 with h171c mutation
120 d1n1ea2
not modelled
97.1
11
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: 6-phosphogluconate dehydrogenase-like, N-terminal domain