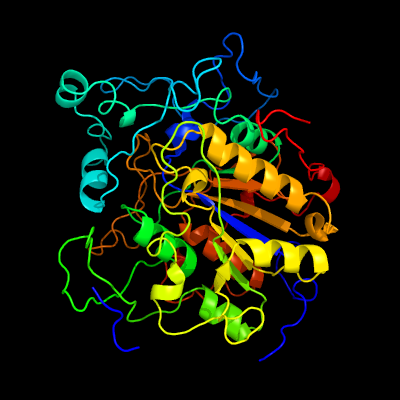
1 c2d1gB_
100.0
22
PDB header: hydrolaseChain: B: PDB Molecule: acid phosphatase;PDBTitle: structure of francisella tularensis acid phosphatase a (acpa) bound to2 orthovanadate
2 c6hr5A_
99.4
15
PDB header: hydrolaseChain: A: PDB Molecule: alpha-l-rhamnosidase/sulfatase (gh78);PDBTitle: structure of the s1_25 family sulfatase module of the rhamnosidase2 fa22250 from formosa agariphila
3 d1auka_
99.4
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
4 c6b1vB_
99.3
16
PDB header: hydrolaseChain: B: PDB Molecule: iota-carrageenan sulfatase;PDBTitle: crystal structure of ps i-cgsb c78s in complex with i-neocarratetraose
5 c4fdiA_
99.3
18
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylgalactosamine-6-sulfatase;PDBTitle: the molecular basis of mucopolysaccharidosis iv a
6 c3lxqB_
99.3
11
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein vp1736;PDBTitle: the crystal structure of a protein in the alkaline phosphatase2 superfamily from vibrio parahaemolyticus to 1.95a
7 c6j66B_
99.3
19
PDB header: hydrolaseChain: B: PDB Molecule: chondroitin sulfate/dermatan sulfate 4-o-endosulfatasePDBTitle: chondroitin sulfate/dermatan sulfate endolytic 4-o-sulfatase
8 d1p49a_
99.2
21
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
9 c3ed4A_
99.2
12
PDB header: transferaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of putative arylsulfatase from escherichia coli
10 c4mivB_
99.2
20
PDB header: hydrolaseChain: B: PDB Molecule: n-sulphoglucosamine sulphohydrolase;PDBTitle: crystal structure of sulfamidase, crystal form l
11 d1fsua_
99.2
18
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
12 c5fqlA_
99.1
16
PDB header: hydrolaseChain: A: PDB Molecule: iduronate-2-sulfatase;PDBTitle: insights into hunter syndrome from the structure of iduronate-2-2 sulfatase
13 c6hhmA_
99.1
15
PDB header: hydrolaseChain: A: PDB Molecule: arylsulfatase;PDBTitle: crystal structure of the family s1_7 ulvan-specific sulfatase fa220702 from formosa agariphila
14 c2qzuA_
99.0
16
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of the putative sulfatase yidj from bacteroides2 fragilis. northeast structural genomics consortium target bfr123
15 c4upkC_
99.0
17
PDB header: hydrolaseChain: C: PDB Molecule: phosphonate monoester hydrolase;PDBTitle: phosphonate monoester hydrolase sppmh from silicibacter pomeroyi
16 c4uopB_
99.0
10
PDB header: transferaseChain: B: PDB Molecule: lipoteichoic acid primase;PDBTitle: crystal structure of the lipoteichoic acid synthase ltap from listeria2 monocytogenes
17 c4ug4H_
99.0
12
PDB header: hydrolaseChain: H: PDB Molecule: choline sulfatase;PDBTitle: crystal structure of a choline sulfatase from sinorhizobium2 melliloti
18 c5g2vA_
99.0
17
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylglucosamine-6-sulfatase;PDBTitle: structure of bt4656 in complex with its substrate d-glucosamine-2-n,2 6-o-disulfate.
19 c2vqrA_
99.0
17
PDB header: hydrolaseChain: A: PDB Molecule: putative sulfatase;PDBTitle: crystal structure of a phosphonate monoester hydrolase from rhizobium2 leguminosarum: a new member of the alkaline phosphatase superfamily
20 c4uorK_
99.0
13
PDB header: transferaseChain: K: PDB Molecule: lipoteichoic acid synthase;PDBTitle: structure of lipoteichoic acid synthase ltas from listeria2 monocytogenes in complex with glycerol phosphate
21 c4uplC_
not modelled
99.0
16
PDB header: hydrolaseChain: C: PDB Molecule: sulfatase family protein;PDBTitle: dimeric sulfatase spas2 from silicibacter pomeroyi
22 c3b5qB_
not modelled
99.0
16
PDB header: hydrolaseChain: B: PDB Molecule: putative sulfatase yidj;PDBTitle: crystal structure of a putative sulfatase (np_810509.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.40 a resolution
23 c4uphA_
not modelled
99.0
18
PDB header: hydrolaseChain: A: PDB Molecule: sulfatase (sulfuric ester hydrolase) protein;PDBTitle: crystal structure of phosphonate monoester hydrolase of agrobacterium2 radiobacter
24 c2w8dB_
not modelled
98.9
11
PDB header: transferaseChain: B: PDB Molecule: processed glycerol phosphate lipoteichoic acid synthase 2;PDBTitle: distinct and essential morphogenic functions for wall- and2 lipo-teichoic acids in bacillus subtilis
25 d1hdha_
not modelled
98.9
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Arylsulfatase
26 c4upiA_
not modelled
98.8
16
PDB header: hydrolaseChain: A: PDB Molecule: sulfatase family protein;PDBTitle: dimeric sulfatase spas1 from silicibacter pomeroyi
27 c5i5fA_
not modelled
98.8
12
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein yejm;PDBTitle: salmonella global domain 191
28 c6a82A_
not modelled
98.7
12
PDB header: transferaseChain: A: PDB Molecule: phosphoethanolamine transferase eptc;PDBTitle: crystal structure of the c-terminal periplasmic domain of eceptc from2 escherichia coli
29 c5k4pA_
not modelled
98.7
16
PDB header: transferaseChain: A: PDB Molecule: probable phosphatidylethanolamine transferase mcr-1;PDBTitle: catalytic domain of mcr-1 phosphoethanolamine transferase
30 c2w5tA_
not modelled
98.6
13
PDB header: transferaseChain: A: PDB Molecule: processed glycerol phosphate lipoteichoic acidPDBTitle: structure-based mechanism of lipoteichoic acid synthesis by2 staphylococcus aureus ltas.
31 c4kayA_
not modelled
98.6
12
PDB header: transferaseChain: A: PDB Molecule: yhbx/yhjw/yijp/yjdb family protein;PDBTitle: structure of the soluble domain of lipooligosaccharide2 phosphoethanolamine transferase a from neisseria meningitidis -3 complex with zn
32 c6bneA_
not modelled
98.5
11
PDB header: transferaseChain: A: PDB Molecule: phosphoethanolamine transferase;PDBTitle: crystal structure of the intrinsic colistin resistance enzyme icr(mc)2 from moraxella catarrhalis, catalytic domain, phosphate-bound complex
33 c5vemA_
not modelled
98.5
11
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 5 (enpp5,2 npp5)
34 c4tn0C_
not modelled
98.4
13
PDB header: transferaseChain: C: PDB Molecule: upf0141 protein yjdb;PDBTitle: crystal structure of the c-terminal periplasmic domain of2 phosphoethanolamine transferase eptc from campylobacter jejuni
35 d1o98a2
not modelled
98.2
12
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: 2,3-Bisphosphoglycerate-independent phosphoglycerate mutase, catalytic domain
36 c5egeD_
not modelled
98.1
12
PDB header: hydrolaseChain: D: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: structure of enpp6, a choline-specific glycerophosphodiester-2 phosphodiesterase
37 c4lqyA_
not modelled
97.9
12
PDB header: hydrolaseChain: A: PDB Molecule: bis(5'-adenosyl)-triphosphatase enpp4;PDBTitle: crystal structure of human enpp4 with amp
38 c5fgnA_
not modelled
97.8
15
PDB header: transferase,hydrolaseChain: A: PDB Molecule: lipooligosaccharide phosphoethanolamine transferase a;PDBTitle: integral membrane protein lipooligosaccharide phosphoethanolamine2 transferase a (epta) from neisseria meningitidis
39 c5u9zB_
not modelled
97.8
9
PDB header: transferaseChain: B: PDB Molecule: phosphoglycerol transferase;PDBTitle: phosphoglycerol transferase gach from streptococcus pyogenes
40 c5udyA_
not modelled
97.7
14
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human alkaline sphingomyelinase (alk-smase, enpp7, npp7)
41 d2i09a1
not modelled
97.7
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: DeoB catalytic domain-like
42 c2gsoB_
not modelled
97.6
14
PDB header: hydrolaseChain: B: PDB Molecule: phosphodiesterase-nucleotide pyrophosphatase;PDBTitle: structure of xac nucleotide pyrophosphatase/phosphodiesterase in2 complex with vanadate
43 c3m8yC_
not modelled
97.4
22
PDB header: isomeraseChain: C: PDB Molecule: phosphopentomutase;PDBTitle: phosphopentomutase from bacillus cereus after glucose-1,6-bisphosphate2 activation
44 c5gz4A_
not modelled
97.0
13
PDB header: hydrolaseChain: A: PDB Molecule: snake venom phosphodiesterase (pde);PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra)
45 c3q3qA_
not modelled
97.0
18
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: crystal structure of spap: an novel alkaline phosphatase from2 bacterium sphingomonas sp. strain bsar-1
46 c5gz5A_
not modelled
96.9
12
PDB header: hydrolaseChain: A: PDB Molecule: snake venom phosphodiesterase (pde);PDBTitle: crystal structure of snake venom phosphodiesterase (pde) from taiwan2 cobra (naja atra atra) in complex with amp
47 c4b56A_
not modelled
96.8
12
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: structure of ectonucleotide pyrophosphatase-phosphodiesterase-12 (npp1)
48 c2i09A_
not modelled
96.4
16
PDB header: isomeraseChain: A: PDB Molecule: phosphopentomutase;PDBTitle: crystal structure of putative phosphopentomutase from streptococcus2 mutans
49 c2zktB_
not modelled
96.3
12
PDB header: isomeraseChain: B: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: structure of ph0037 protein from pyrococcus horikoshii
50 c6c02B_
not modelled
96.2
10
PDB header: hydrolaseChain: B: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: human ectonucleotide pyrophosphatase / phosphodiesterase 3 (enpp3,2 npp3, cd203c), inactive (t205a), n594s, with alpha,beta-methylene-atp3 (ampcpp)
51 c2xr9A_
not modelled
94.8
15
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2)
52 c2xrgA_
not modelled
94.4
14
PDB header: hydrolaseChain: A: PDB Molecule: ectonucleotide pyrophosphatase/phosphodiesterase familyPDBTitle: crystal structure of autotaxin (enpp2) in complex with the2 ha155 boronic acid inhibitor
53 c5kgmA_
not modelled
93.4
25
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: 2.95a resolution structure of apo independent phosphoglycerate mutase2 from c. elegans (monoclinic form)
54 c2e76D_
not modelled
92.7
15
PDB header: photosynthesisChain: D: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
55 c3igzB_
not modelled
90.5
14
PDB header: isomeraseChain: B: PDB Molecule: cofactor-independent phosphoglycerate mutase;PDBTitle: crystal structures of leishmania mexicana phosphoglycerate2 mutase at low cobalt concentration
56 c5tj3A_
not modelled
88.0
10
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase pafa;PDBTitle: crystal structure of wild type alkaline phosphatase pafa to 1.7a2 resolution
57 c1o98A_
not modelled
86.7
20
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independentPDBTitle: 1.4a crystal structure of phosphoglycerate mutase from2 bacillus stearothermophilus complexed with3 2-phosphoglycerate
58 c2fynO_
not modelled
85.3
22
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
59 d1ei6a_
not modelled
78.7
13
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Phosphonoacetate hydrolase
60 c2pq4B_
not modelled
75.4
33
PDB header: chaperone/oxidoreductaseChain: B: PDB Molecule: periplasmic nitrate reductase precursor;PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
61 c2fyuE_
not modelled
74.5
12
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit,PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
62 c5vpuA_
not modelled
73.4
15
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: crystal structure of 2,3-bisphosphoglycerate-independent2 phosphoglycerate mutase bound to 3-phosphoglycerate, from3 acinetobacter baumannii
63 c1p84E_
not modelled
73.1
14
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
64 c4my4A_
not modelled
72.4
22
PDB header: isomeraseChain: A: PDB Molecule: 2,3-bisphosphoglycerate-independent phosphoglyceratePDBTitle: crystal structure of phosphoglycerate mutase from staphylococcus2 aureus.
65 d1y6va1
not modelled
69.8
17
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase
66 c6fo2R_
not modelled
65.5
14
PDB header: membrane proteinChain: R: PDB Molecule: cytochrome b-c1 complex subunit rieske, mitochondrial;PDBTitle: cryoem structure of bovine cytochrome bc1 with no ligand bound
67 c3szzA_
not modelled
63.0
18
PDB header: hydrolaseChain: A: PDB Molecule: phosphonoacetate hydrolase;PDBTitle: crystal structure of phosphonoacetate hydrolase from sinorhizobium2 meliloti 1021 in complex with acetate
68 c2w0yB_
not modelled
61.3
31
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
69 c3couA_
not modelled
41.8
27
PDB header: hydrolaseChain: A: PDB Molecule: nucleoside diphosphate-linked moiety x motif 16;PDBTitle: crystal structure of human nudix motif 16 (nudt16)
70 d1k0ma2
not modelled
40.1
23
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
71 d1okta2
not modelled
38.7
17
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
72 c3e2dB_
not modelled
28.9
27
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active vibrio sp.2 alkaline phosphatase
73 c2iucB_
not modelled
28.2
14
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic bacterium tab5
74 c2x98A_
not modelled
27.1
30
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
75 d2gsra2
not modelled
25.4
21
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
76 c3wbhB_
not modelled
25.2
21
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structural characteristics of alkaline phosphatase from a moderately2 halophilic bacteria halomonas sp.593
77 c4ke3D_
not modelled
24.5
26
PDB header: transferaseChain: D: PDB Molecule: glutathione s-transferase domain;PDBTitle: crystal structure of a glutathione transferase family member from2 burkholderia graminis, target efi-507264, no gsh, disordered domains,3 space group p21, form(2)
78 d1g57a_
not modelled
24.0
29
Fold: YrdC/RibBSuperfamily: YrdC/RibBFamily: 3,4-dihydroxy-2-butanone 4-phosphate synthase, DHBP synthase, RibB
79 c3kvhA_
not modelled
23.6
31
PDB header: rna binding proteinChain: A: PDB Molecule: protein syndesmos;PDBTitle: crystal structure of human protein syndesmos (nudt16-like protein)
80 c5jx6C_
not modelled
23.4
14
PDB header: hydrolaseChain: C: PDB Molecule: glucanase;PDBTitle: gh6 orpinomyces sp. y102 enzyme
81 d2e74d2
not modelled
22.5
22
Fold: Single transmembrane helixSuperfamily: ISP transmembrane anchorFamily: ISP transmembrane anchor
82 d1dysa_
not modelled
22.0
14
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
83 c6jcnB_
not modelled
21.9
11
PDB header: transferaseChain: B: PDB Molecule: dehydrodolichyl diphosphate synthase complex subunit nus1;PDBTitle: yeast dehydrodolichyl diphosphate synthase complex subunit nus1
84 c6acsA_
not modelled
20.5
27
PDB header: transferaseChain: A: PDB Molecule: ditrans,polycis-undecaprenyl-diphosphate synthase ((2e,6e)-PDBTitle: poly-cis-prenyltransferase
85 c1x5eA_
not modelled
19.7
13
PDB header: electron transportChain: A: PDB Molecule: thioredoxin domain containing protein 1;PDBTitle: the solution structure of the thioredoxin-like domain of2 human thioredoxin-related transmembrane protein
86 d1qjwa_
not modelled
19.5
23
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
87 c5j43F_
not modelled
19.1
56
PDB header: toxinChain: F: PDB Molecule: trna nuclease cdia;PDBTitle: cdia-ct from uropathogenic escherichia coli in complex with cysk
88 d1v2aa2
not modelled
18.7
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
89 d1b5ea_
not modelled
18.5
17
Fold: Thymidylate synthase/dCMP hydroxymethylaseSuperfamily: Thymidylate synthase/dCMP hydroxymethylaseFamily: Thymidylate synthase/dCMP hydroxymethylase
90 d1m0ua2
not modelled
17.8
11
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
91 c3a64A_
not modelled
17.4
14
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: crystal structure of cccel6c, a glycoside hydrolase family 62 enzyme, from coprinopsis cinerea
92 d1uoza_
not modelled
17.2
9
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
93 c4yn5A_
not modelled
16.9
11
PDB header: hydrolaseChain: A: PDB Molecule: mannan endo-1,4-beta-mannosidase;PDBTitle: catalytic domain of bacillus sp. jamb-750 gh26 endo-beta-1,4-mannanase
94 c4nv4A_
not modelled
16.3
33
PDB header: hydrolaseChain: A: PDB Molecule: signal peptidase i;PDBTitle: 1.8 angstrom crystal structure of signal peptidase i from bacillus2 anthracis.
95 c5xczA_
not modelled
15.9
17
PDB header: hydrolaseChain: A: PDB Molecule: glucanase;PDBTitle: structure of the cellobiohydrolase cel6a from phanerochaete2 chrysosporium in complex with cellobiose at 2.1 angstrom
96 c6hwhB_
not modelled
15.3
26
PDB header: electron transportChain: B: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
97 c1q7tA_
not modelled
15.2
27
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein rv1170;PDBTitle: rv1170 (mshb) from mycobacterium tuberculosis
98 d2gsqa2
not modelled
15.1
15
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Glutathione S-transferase (GST), N-terminal domain
99 d1txka2
not modelled
14.8
36
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: MdoG-like