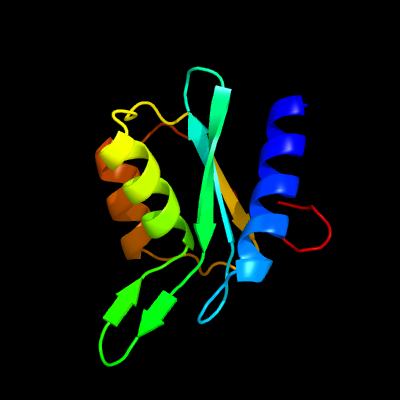
| 1 | d2fr5a1
|
|
 |
100.0 |
22 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
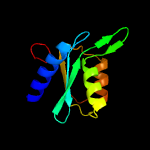
| 2 | d2z3ga1
|
|
 |
100.0 |
23 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
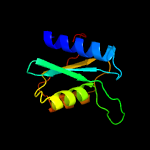
| 3 | d1mq0a_
|
|
 |
100.0 |
21 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
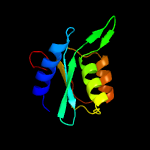
| 4 | c3oj6C_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:blasticidin-s deaminase;
PDBTitle: crystal structure of blasticidin s deaminase from coccidioides immitis
|
|
|
|
| 5 | d1uwza_
|
|
 |
100.0 |
27 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
| 6 | c3b8fB_
|
|
 |
100.0 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative blasticidin s deaminase;
PDBTitle: crystal structure of the cytidine deaminase from bacillus anthracis
|
|
|
|
| 7 | d2d30a1
|
|
 |
100.0 |
23 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
| 8 | c3dmoD_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: D: PDB Molecule:cytidine deaminase;
PDBTitle: 1.6 a crystal structure of cytidine deaminase from burkholderia2 pseudomallei
|
|
|
|
| 9 | c3r2nC_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: C: PDB Molecule:cytidine deaminase;
PDBTitle: crystal structure of cytidine deaminase from mycobacterium leprae
|
|
|
|
| 10 | d1r5ta_
|
|
 |
100.0 |
13 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
| 11 | c3ijfX_
|
|
 |
100.0 |
29 |
PDB header:hydrolase
Chain: X: PDB Molecule:cytidine deaminase;
PDBTitle: crystal structure of cytidine deaminase from mycobacterium2 tuberculosis
|
|
|
|
| 12 | c4eg2G_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: G: PDB Molecule:cytidine deaminase;
PDBTitle: 2.2 angstrom crystal structure of cytidine deaminase from vibrio2 cholerae in complex with zinc and uridine
|
|
|
|
| 13 | c1alnA_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytidine deaminase;
PDBTitle: crystal structure of cytidine deaminase complexed with 3-deazacytidine
|
|
|
|
| 14 | d1alna1
|
|
 |
99.9 |
26 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
| 15 | d1alna2
|
|
 |
99.9 |
24 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Cytidine deaminase |
|
|
|
| 16 | c4p9eA_
|
|
 |
98.4 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: crystal structure of dcmp deaminase from the cyanophage s-tim5 in apo2 form
|
|
|
|
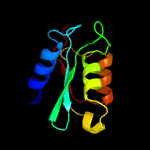
| 17 | d1p6oa_
|
|
 |
98.0 |
12 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
|
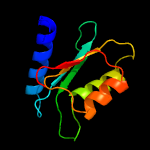
| 18 | c5xkrA_
|
|
 |
97.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:cmp/dcmp deaminase, zinc-binding protein;
PDBTitle: crystal structure of msmeg3575 in complex with benzoguanamine
|
|
|
|
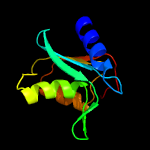
| 19 | c2hvwC_
|
|
 |
97.9 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: crystal structure of dcmp deaminase from streptococcus mutans
|
|
|
|
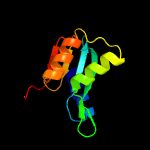
| 20 | c2w4lC_
|
|
 |
97.8 |
14 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxycytidylate deaminase;
PDBTitle: human dcmp deaminase
|
|
|
|
| 21 | c5jfyC_ |
|
not modelled |
97.7 |
9 |
PDB header:hydrolase
Chain: C: PDB Molecule:deoxycytidine deaminase;
PDBTitle: crystal structure of a plant cytidine deaminase
|
|
|
| 22 | d2b3ja1 |
|
not modelled |
97.6 |
14 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 23 | d2g84a1 |
|
not modelled |
97.4 |
17 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 24 | c2hxvA_ |
|
not modelled |
97.4 |
24 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:diaminohydroxyphosphoribosylaminopyrimidine deaminase/ 5-
PDBTitle: crystal structure of a diaminohydroxyphosphoribosylaminopyrimidine2 deaminase/ 5-amino-6-(5-phosphoribosylamino)uracil reductase (tm1828)3 from thermotoga maritima at 1.80 a resolution
|
|
|
| 25 | d1z3aa1 |
|
not modelled |
97.3 |
13 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 26 | d1wkqa_ |
|
not modelled |
97.3 |
12 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 27 | c3ocqA_ |
|
not modelled |
97.1 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative cytosine/adenosine deaminase;
PDBTitle: crystal structure of trna-specific adenosine deaminase from salmonella2 enterica
|
|
|
| 28 | d1wwra1 |
|
not modelled |
97.1 |
15 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 29 | c2o7pA_ |
|
not modelled |
97.1 |
15 |
PDB header:hydrolase, oxidoreductase
Chain: A: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: the crystal structure of ribd from escherichia coli in complex with2 the oxidised nadp+ cofactor in the active site of the reductase3 domain
|
|
|
| 30 | c2nx8A_ |
|
not modelled |
97.1 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:trna-specific adenosine deaminase;
PDBTitle: the crystal structure of the trna-specific adenosine deaminase from2 streptococcus pyogenes
|
|
|
| 31 | d1vq2a_ |
|
not modelled |
97.0 |
15 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 32 | d2a8na1 |
|
not modelled |
97.0 |
15 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 33 | c2d5nB_ |
|
not modelled |
95.8 |
24 |
PDB header:hydrolase, oxidoreductase
Chain: B: PDB Molecule:riboflavin biosynthesis protein ribd;
PDBTitle: crystal structure of a bifunctional deaminase and reductase2 involved in riboflavin biosynthesis
|
|
|
| 34 | d2b3za2 |
|
not modelled |
95.6 |
18 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 35 | c3dh1D_ |
|
not modelled |
95.2 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:trna-specific adenosine deaminase 2;
PDBTitle: crystal structure of human trna-specific adenosine-34 deaminase2 subunit adat2
|
|
|
| 36 | c3zpgA_ |
|
not modelled |
95.2 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:ribd;
PDBTitle: acinetobacter baumannii ribd, form 2
|
|
|
| 37 | d2hxva2 |
|
not modelled |
94.9 |
17 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Deoxycytidylate deaminase-like |
|
|
| 38 | d1l5ja3 |
|
not modelled |
39.6 |
9 |
Fold:Aconitase iron-sulfur domain
Superfamily:Aconitase iron-sulfur domain
Family:Aconitase iron-sulfur domain |
|
|
| 39 | c4uwqK_ |
|
not modelled |
33.5 |
17 |
PDB header:hydrolase
Chain: K: PDB Molecule:soxy protein;
PDBTitle: crystal structure of the disulfide-linked complex of the2 thiosulfodyrolase soxb with the carrier-protein soxyz from3 thermus thermophilus
|
|
|
| 40 | c3dmlA_ |
|
not modelled |
32.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the periplasmic thioredoxin soxs from paracoccus2 pantotrophus (reduced form)
|
|
|
| 41 | c2zkqn_ |
|
not modelled |
30.8 |
17 |
PDB header:ribosomal protein/rna
Chain: N: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
|
|
|
| 42 | c1l5jB_ |
|
not modelled |
30.8 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:aconitate hydratase 2;
PDBTitle: crystal structure of e. coli aconitase b.
|
|
|
| 43 | c5xyid_ |
|
not modelled |
29.3 |
25 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein s3, putative;
PDBTitle: small subunit of trichomonas vaginalis ribosome
|
|
|
| 44 | c3c4nB_ |
|
not modelled |
21.1 |
23 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein dr_0571;
PDBTitle: crystal structure of dr_0571 protein from deinococcus radiodurans in2 complex with adp. northeast structural genomics consortium target3 drr125
|
|
|
| 45 | d1njra_ |
|
not modelled |
20.1 |
11 |
Fold:Macro domain-like
Superfamily:Macro domain-like
Family:Macro domain |
|
|
| 46 | c3g9bA_ |
|
not modelled |
20.0 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-diphosphooligosaccharide-protein
PDBTitle: crystal structure of reduced ost6l
|
|
|
| 47 | d3d37a2 |
|
not modelled |
18.9 |
0 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
|
|
| 48 | d1hyua4 |
|
not modelled |
18.5 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:PDI-like |
|
|
| 49 | c3zey8_ |
|
not modelled |
18.1 |
25 |
PDB header:ribosome
Chain: 8: PDB Molecule:ribosomal protein s29, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 50 | c2xznN_ |
|
not modelled |
17.8 |
33 |
PDB header:ribosome
Chain: N: PDB Molecule:rps29e;
PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 2
|
|
|
| 51 | c3j20P_ |
|
not modelled |
17.5 |
33 |
PDB header:ribosome
Chain: P: PDB Molecule:30s ribosomal protein s14p type z;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
|
|
|
| 52 | c2oxgD_ |
|
not modelled |
17.5 |
22 |
PDB header:transport protein
Chain: D: PDB Molecule:soxy protein;
PDBTitle: the soxyz complex of paracoccus pantotrophus
|
|
|
| 53 | d1b4ub_ |
|
not modelled |
17.0 |
14 |
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
|
|
| 54 | c3jyvN_ |
|
not modelled |
16.7 |
25 |
PDB header:ribosome
Chain: N: PDB Molecule:40s ribosomal protein s29(a);
PDBTitle: structure of the 40s rrna and proteins and p/e trna for eukaryotic2 ribosome based on cryo-em map of thermomyces lanuginosus ribosome at3 8.9a resolution
|
|
|
| 55 | c5cb9A_ |
|
not modelled |
15.7 |
29 |
PDB header:lyase
Chain: A: PDB Molecule:glyoxalase/bleomycin resistance protein/dioxygenase;
PDBTitle: crystal structure of c-as lyase with mercaptoethonal
|
|
|
| 56 | d1liua3 |
|
not modelled |
15.5 |
20 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 57 | c3wrbB_ |
|
not modelled |
15.1 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gallate dioxygenase;
PDBTitle: crystal structure of the anaerobic h124f desb-gallate complex
|
|
|
| 58 | c5xxud_ |
|
not modelled |
15.1 |
25 |
PDB header:ribosome
Chain: D: PDB Molecule:ribosomal protein us3;
PDBTitle: small subunit of toxoplasma gondii ribosome
|
|
|
| 59 | d1booa_ |
|
not modelled |
14.2 |
5 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Type II DNA methylase |
|
|
| 60 | c1oatB_ |
|
not modelled |
14.2 |
17 |
PDB header:aminotransferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: ornithine aminotransferase
|
|
|
| 61 | c3bh1A_ |
|
not modelled |
13.3 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0371 protein dip2346;
PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
|
|
|
| 62 | c6az1S_ |
|
not modelled |
12.9 |
25 |
PDB header:ribosome/antibiotic
Chain: S: PDB Molecule:ribosomal protein s14;
PDBTitle: cryo-em structure of the small subunit of leishmania ribosome bound to2 paromomycin
|
|
|
| 63 | d1e0ta3 |
|
not modelled |
12.8 |
26 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 64 | d2g50a3 |
|
not modelled |
12.6 |
24 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 65 | d3bzka5 |
|
not modelled |
12.5 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Tex RuvX-like domain-like |
|
|
| 66 | d1pkla3 |
|
not modelled |
12.3 |
17 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 67 | c1s1hN_ |
|
not modelled |
11.8 |
27 |
PDB header:ribosome
Chain: N: PDB Molecule:40s ribosomal protein s29-b;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
|
|
|
| 68 | c6h9cD_ |
|
not modelled |
11.6 |
59 |
PDB header:virus
Chain: D: PDB Molecule:vp7;
PDBTitle: cryo-em structure of archaeal extremophilic internal membrane-2 containing haloarcula californiae icosahedral virus 1 (hciv-1) at3 3.74 angstroms resolution.
|
|
|
| 69 | c4pv3D_ |
|
not modelled |
11.6 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:l-asparaginase beta subunit;
PDBTitle: crystal structure of potassium-dependent plant-type l-asparaginase2 from phaseolus vulgaris in complex with na+ cations
|
|
|
| 70 | d1siqa2 |
|
not modelled |
11.4 |
22 |
Fold:Acyl-CoA dehydrogenase NM domain-like
Superfamily:Acyl-CoA dehydrogenase NM domain-like
Family:Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains |
|
|
| 71 | c6ib5B_ |
|
not modelled |
11.4 |
24 |
PDB header:flavoprotein
Chain: B: PDB Molecule:tryptophan 6-halogenase;
PDBTitle: mutant of flavin-dependent tryptophan halogenase thal with altered2 regioselectivity (thal-rebh5)
|
|
|
| 72 | c6bwsA_ |
|
not modelled |
11.3 |
22 |
PDB header:unknown function
Chain: A: PDB Molecule:glycolate utilization protein;
PDBTitle: crystal structure of efga from methylobacterium extorquens
|
|
|
| 73 | d1gawa1 |
|
not modelled |
10.6 |
19 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 74 | c5n1tA_ |
|
not modelled |
10.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:flavin-binding subunit of sulfide dehydrogenase;
PDBTitle: crystal structure of complex between flavocytochrome c and copper2 chaperone copc from t. paradoxus
|
|
|
| 75 | c3vowB_ |
|
not modelled |
10.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable dna dc->du-editing enzyme apobec-3c;
PDBTitle: crystal structure of the human apobec3c having hiv-1 vif-binding2 interface
|
|
|
| 76 | d1fnda1 |
|
not modelled |
10.2 |
19 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 77 | c2mzzA_ |
|
not modelled |
10.2 |
6 |
PDB header:hydrolase, antiviral protein
Chain: A: PDB Molecule:apolipoprotein b mrna-editing enzyme, catalytic
PDBTitle: nmr structure of apobec3g ntd variant, sntd
|
|
|
| 78 | c1senA_ |
|
not modelled |
9.8 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:thioredoxin-like protein p19;
PDBTitle: endoplasmic reticulum protein rp19 o95881
|
|
|
| 79 | d1sena_ |
|
not modelled |
9.8 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
|
|
| 80 | c4yodA_ |
|
not modelled |
9.1 |
6 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin-like protein;
PDBTitle: crystal structure of a thioredoxin-like protein (baccac_02376) from2 bacteroides caccae atcc 43185 at 1.90 a resolution
|
|
|
| 81 | c4ml1D_ |
|
not modelled |
9.1 |
5 |
PDB header:isomerase
Chain: D: PDB Molecule:dsbp;
PDBTitle: disulfide isomerase (dsbp) from multidrug resistance inca/c2 transferable plasmid in oxidized state (p212121 space group)
|
|
|
| 82 | c2e7uA_ |
|
not modelled |
8.4 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate-1-semialdehyde 2,1-aminomutase;
PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-aminomutase from2 thermus thermophilus hb8
|
|
|
| 83 | c4e6kI_ |
|
not modelled |
8.3 |
11 |
PDB header:metal binding protein/electron transport
Chain: I: PDB Molecule:bacterioferritin-associated ferredoxin;
PDBTitle: 2.0 a resolution structure of pseudomonas aeruginosa bacterioferritin2 (bfrb) in complex with bacterioferritin associated ferredoxin (bfd)
|
|
|
| 84 | c3g12A_ |
|
not modelled |
8.3 |
36 |
PDB header:lyase
Chain: A: PDB Molecule:putative lactoylglutathione lyase;
PDBTitle: crystal structure of a putative lactoylglutathione lyase from2 bdellovibrio bacteriovorus
|
|
|
| 85 | c2fgxA_ |
|
not modelled |
8.2 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative thioredoxin;
PDBTitle: solution nmr structure of protein ne2328 from nitrosomonas2 europaea. northeast structural genomics consortium target3 net3.
|
|
|
| 86 | c4kp1A_ |
|
not modelled |
8.2 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:isopropylmalate/citramalate isomerase large subunit;
PDBTitle: crystal structure of ipm isomerase large subunit from methanococcus2 jannaschii (mj0499)
|
|
|
| 87 | c4mubA_ |
|
not modelled |
8.1 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:sulfotransferase;
PDBTitle: schistosoma mansoni (blood fluke) sulfotransferase/oxamniquine complex
|
|
|
| 88 | c1yspA_ |
|
not modelled |
7.9 |
14 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator kdgr;
PDBTitle: crystal structure of the c-terminal domain of e. coli transcriptional2 regulator kdgr.
|
|
|
| 89 | c3ruyB_ |
|
not modelled |
7.8 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:ornithine aminotransferase;
PDBTitle: crystal structure of the ornithine-oxo acid transaminase rocd from2 bacillus anthracis
|
|
|
| 90 | d1a3xa3 |
|
not modelled |
7.8 |
22 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:Pyruvate kinase, C-terminal domain |
|
|
| 91 | c4z04A_ |
|
not modelled |
7.7 |
27 |
PDB header:lyase
Chain: A: PDB Molecule:glyoxalase/bleomycin resistance /dioxygenase superfamily
PDBTitle: crystal structure of a probable lactoylglutathione lyase from brucella2 melitensis in complex with glutathione
|
|
|
| 92 | c3fosA_ |
|
not modelled |
7.7 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:sensor protein;
PDBTitle: crystal structure of two-component sensor histidine kinase domain from2 bacillus subtilis subsp. subtilis str. 168
|
|
|
| 93 | c4m90A_ |
|
not modelled |
7.6 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:tumor suppressor candidate 3;
PDBTitle: crystal structure of oxidized hn33/tusc3
|
|
|
| 94 | d2bmwa1 |
|
not modelled |
7.6 |
15 |
Fold:Reductase/isomerase/elongation factor common domain
Superfamily:Riboflavin synthase domain-like
Family:Ferredoxin reductase FAD-binding domain-like |
|
|
| 95 | d1g5ca_ |
|
not modelled |
7.5 |
9 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 96 | c2k8vA_ |
|
not modelled |
7.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:thioredoxin domain-containing protein 12;
PDBTitle: solution structure of oxidised erp18
|
|
|
| 97 | c5k83C_ |
|
not modelled |
7.3 |
9 |
PDB header:hydrolase
Chain: C: PDB Molecule:apolipoprotein b mrna editing enzyme, catalytic peptide-
PDBTitle: crystal structure of a primate apobec3g n-domain, in complex with2 ssdna
|
|
|
| 98 | c4e6kG_ |
|
not modelled |
7.3 |
11 |
PDB header:metal binding protein/electron transport
Chain: G: PDB Molecule:bacterioferritin-associated ferredoxin;
PDBTitle: 2.0 a resolution structure of pseudomonas aeruginosa bacterioferritin2 (bfrb) in complex with bacterioferritin associated ferredoxin (bfd)
|
|
|
| 99 | d2f9wa1 |
|
not modelled |
7.2 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:CoaX-like |
|
|