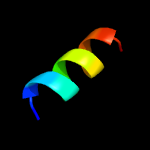
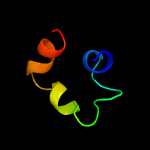
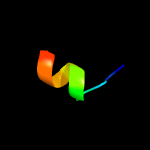
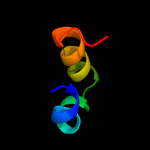
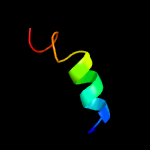
1 d1o82a_
27.9
41
Fold: Saposin-likeSuperfamily: Bacteriocin AS-48Family: Bacteriocin AS-48
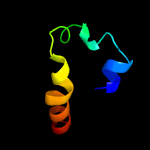
2 c5kw2A_
21.2
16
PDB header: fatty acid binding protein/hydrolaseChain: A: PDB Molecule: free fatty acid receptor 1,lysozyme,free fatty acidPDBTitle: the extra-helical binding site of gpr40 and the structural basis for2 allosteric agonism and incretin stimulation
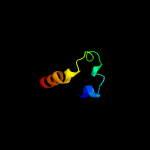
3 c2kk1A_
20.7
9
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase abl2;PDBTitle: solution structure of c-terminal domain of tyrosine-protein2 kinase abl2 from homo sapiens, northeast structural3 genomics consortium (nesg) target hr5537a
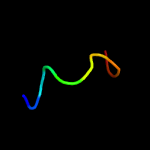
4 c1iijA_
16.3
33
PDB header: signaling proteinChain: A: PDB Molecule: erbb-2 receptor protein-tyrosine kinase;PDBTitle: solution structure of the neu/erbb-2 membrane spanning2 segment
5 d1jmxa2
16.3
22
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
6 d1pbya2
15.0
24
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
7 c2mmjA_
11.9
40
PDB header: antimicrobial proteinChain: A: PDB Molecule: maculatin g15;PDBTitle: structure of a peptoid analogue of maculatin g15 in dpc micelles
8 c4wrpB_
11.8
17
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: the c-terminal domain of gene product lpg0944 from legionella2 pneumophila subsp. pneumophila str. philadelphia 1
9 c1jmxA_
10.4
24
PDB header: oxidoreductaseChain: A: PDB Molecule: amine dehydrogenase;PDBTitle: crystal structure of a quinohemoprotein amine dehydrogenase2 from pseudomonas putida
10 c5lc5c_
9.1
9
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class2
11 c2ld7B_
8.3
12
PDB header: transcriptionChain: B: PDB Molecule: paired amphipathic helix protein sin3a;PDBTitle: solution structure of the msin3a pah3-sap30 sid complex
12 c2l53B_
7.9
29
PDB header: ca-binding protein/proton transportChain: B: PDB Molecule: voltage-gated sodium channel type v alpha isoform bPDBTitle: solution nmr structure of apo-calmodulin in complex with the iq motif2 of human cardiac sodium channel nav1.5
13 c2kpeB_
7.2
40
PDB header: membrane proteinChain: B: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
14 c2kpeA_
7.2
40
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
15 c2amnA_
7.1
21
PDB header: antimicrobial proteinChain: A: PDB Molecule: cathelicidin;PDBTitle: solution structure of fowlicidin-1, a novel cathelicidin2 antimicrobial peptide from chicken
16 c5eh6A_
6.9
36
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane monomer in2 lipidic cubic phase
17 c5eh4A_
6.9
36
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
18 c5eh4B_
6.9
36
PDB header: membrane proteinChain: B: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
19 c5eh4D_
6.9
36
PDB header: membrane proteinChain: D: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
20 c5eh4C_
6.9
36
PDB header: membrane proteinChain: C: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
21 c6ca8A_
not modelled
6.8
13
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase 2;PDBTitle: crystal structure of plasmodium falciparum topoisomerase ii dna-2 binding, cleavage and re-ligation domain
22 c5ldwc_
not modelled
6.3
9
PDB header: oxidoreductaseChain: C: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 3,PDBTitle: structure of mammalian respiratory complex i, class1
23 c3j47R_
not modelled
5.6
36
PDB header: protein bindingChain: R: PDB Molecule: 26s proteasome regulatory subunit rpn7;PDBTitle: formation of an intricate helical bundle dictates the assembly of the2 26s proteasome lid
24 c3ixzB_
not modelled
5.5
28
PDB header: hydrolaseChain: B: PDB Molecule: potassium-transporting atpase subunit beta;PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
25 c4oojB_
not modelled
5.2
27
PDB header: unknown functionChain: B: PDB Molecule: sidc, interaptin;PDBTitle: crystal structure of the n-terminal domain of the legionella2 pneumophila protein sidc at 2.4a resolution