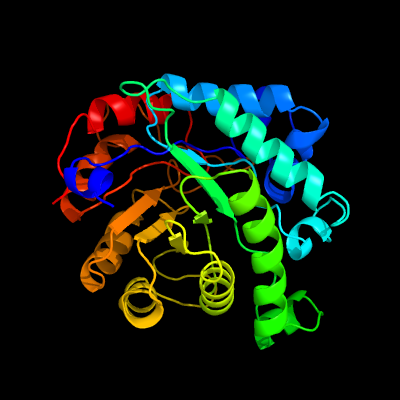
| 1 | d3bofa2
|
|
 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
|
|
|
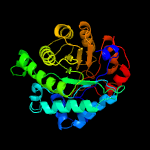
| 2 | c3bolB_
|
|
 |
100.0 |
26 |
PDB header:transferase
Chain: B: PDB Molecule:5-methyltetrahydrofolate s-homocysteine
PDBTitle: cobalamin-dependent methionine synthase (1-566) from2 thermotoga maritima complexed with zn2+
|
|
|
|
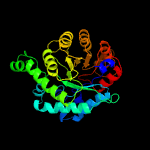
| 3 | c4cczA_
|
|
 |
100.0 |
27 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: crystal structure of human 5-methyltetrahydrofolate-homocysteine2 methyltransferase, the homocysteine and folate binding domains
|
|
|
|
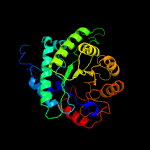
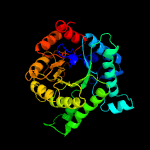
| 4 | d1umya_
|
|
 |
100.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
|
|
|
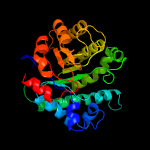
| 5 | c5dmnA_
|
|
 |
100.0 |
47 |
PDB header:transferase
Chain: A: PDB Molecule:homocysteine s-methyltransferase;
PDBTitle: crystal structure of the homocysteine methyltransferase mmum from2 escherichia coli, apo form
|
|
|
|
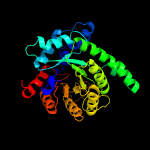
| 6 | d1lt7a_
|
|
 |
100.0 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Homocysteine S-methyltransferase
Family:Homocysteine S-methyltransferase |
|
|
|
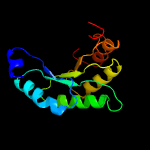
| 7 | c5ks8D_
|
|
 |
96.0 |
13 |
PDB header:ligase
Chain: D: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
|
| 8 | c4mwaA_
|
|
 |
95.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
|
| 9 | c3igsB_
|
|
 |
94.8 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
|
|
|
| 10 | c2nx9B_
|
|
 |
94.7 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:oxaloacetate decarboxylase 2, subunit alpha;
PDBTitle: crystal structure of the carboxyltransferase domain of the2 oxaloacetate decarboxylase na+ pump from vibrio cholerae
|
|
|
|
| 11 | d1muma_
|
|
 |
93.5 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
|
| 12 | c3eooL_
|
|
 |
91.9 |
21 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
|
| 13 | c2dwuA_
|
|
 |
91.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase isoform race1 from bacillus2 anthracis
|
|
|
|
| 14 | c3vndD_
|
|
 |
91.6 |
12 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
|
| 15 | c2ekcA_
|
|
 |
90.5 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
|
| 16 | c1zfjA_
|
|
 |
90.5 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
|
|
|
| 17 | d2fdsa1
|
|
 |
90.3 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
|
| 18 | c2fdsA_
|
|
 |
90.3 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:orotidine-monophosphate-decarboxylase;
PDBTitle: crystal structure of plasmodium berghei orotidine 5'-monophosphate2 decarboxylase (ortholog of plasmodium falciparum pf10_0225)
|
|
|
|
| 19 | d2ffca1
|
|
 |
89.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
|
| 20 | c1rr2A_
|
|
 |
88.7 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:transcarboxylase 5s subunit;
PDBTitle: propionibacterium shermanii transcarboxylase 5s subunit bound to 2-2 ketobutyric acid
|
|
|
|
| 21 | c2infB_ |
|
not modelled |
88.2 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from bacillus2 subtilis
|
|
|
| 22 | d1j93a_ |
|
not modelled |
88.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
| 23 | c5ey5A_ |
|
not modelled |
87.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:lbcats-a;
PDBTitle: lbcats
|
|
|
| 24 | c3cyvA_ |
|
not modelled |
87.4 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 shigella flexineri: new insights into its catalytic3 mechanism
|
|
|
| 25 | c3ih1A_ |
|
not modelled |
87.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 26 | c3hfrA_ |
|
not modelled |
87.1 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of glutamate racemase from listeria monocytogenes
|
|
|
| 27 | c3n2xB_ |
|
not modelled |
87.1 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:uncharacterized protein yage;
PDBTitle: crystal structure of yage, a prophage protein belonging to the2 dihydrodipicolinic acid synthase family from e. coli k12 in complex3 with pyruvate
|
|
|
| 28 | c5xzoA_ |
|
not modelled |
86.9 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-xylanase;
PDBTitle: crystal structure of gh10 xylanase xyl10c from bispora. sp mey-1
|
|
|
| 29 | d1wbha1 |
|
not modelled |
86.8 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 30 | c5zknA_ |
|
not modelled |
86.6 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate 2-epimerase from2 fusobacterium nucleatum
|
|
|
| 31 | c1zlpA_ |
|
not modelled |
86.5 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 32 | d1a3xa2 |
|
not modelled |
86.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 33 | c4zr8B_ |
|
not modelled |
86.0 |
24 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: structure of uroporphyrinogen decarboxylase from acinetobacter2 baumannii
|
|
|
| 34 | c4mg4G_ |
|
not modelled |
85.9 |
18 |
PDB header:unknown function
Chain: G: PDB Molecule:phosphonomutase;
PDBTitle: crystal structure of a putative phosphonomutase from burkholderia2 cenocepacia j2315
|
|
|
| 35 | c4axkB_ |
|
not modelled |
85.9 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:1-(5-phosphoribosyl)-5-((5'-phosphoribosylamino)
PDBTitle: crystal structure of subhisa from the thermophile corynebacterium2 efficiens
|
|
|
| 36 | c4i7vD_ |
|
not modelled |
85.7 |
19 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: agrobacterium tumefaciens dhdps with pyruvate
|
|
|
| 37 | c3q58A_ |
|
not modelled |
85.5 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
|
|
| 38 | c4jn6C_ |
|
not modelled |
84.8 |
19 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal structure of the aldolase-dehydrogenase complex from2 mycobacterium tuberculosis hrv37
|
|
|
| 39 | d1xcfa_ |
|
not modelled |
84.2 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 40 | c3pm6B_ |
|
not modelled |
84.0 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:putative fructose-bisphosphate aldolase;
PDBTitle: crystal structure of a putative fructose-1,6-biphosphate aldolase from2 coccidioides immitis solved by combined sad mr
|
|
|
| 41 | c3lyeA_ |
|
not modelled |
83.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 42 | c3fa4D_ |
|
not modelled |
83.8 |
19 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 43 | c4x2rA_ |
|
not modelled |
83.7 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of pria from actinomyces urogenitalis
|
|
|
| 44 | c3r89A_ |
|
not modelled |
83.5 |
7 |
PDB header:lyase
Chain: A: PDB Molecule:orotidine 5'-phosphate decarboxylase;
PDBTitle: crystal structure of orotidine 5-phosphate decarboxylase from2 anaerococcus prevotii dsm 20548
|
|
|
| 45 | d2q8za1 |
|
not modelled |
83.5 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 46 | d1m3ua_ |
|
not modelled |
83.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 47 | c3fkkA_ |
|
not modelled |
83.1 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:l-2-keto-3-deoxyarabonate dehydratase;
PDBTitle: structure of l-2-keto-3-deoxyarabonate dehydratase
|
|
|
| 48 | c4exqA_ |
|
not modelled |
81.7 |
19 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase (upd) from2 burkholderia thailandensis e264
|
|
|
| 49 | c2ze3A_ |
|
not modelled |
81.2 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 50 | c3b8iF_ |
|
not modelled |
81.0 |
21 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 51 | d1gvfa_ |
|
not modelled |
80.4 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 52 | c2jfoB_ |
|
not modelled |
78.8 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of enterococcus faecalis glutamate2 racemase in complex with d- and l-glutamate
|
|
|
| 53 | c4fxsA_ |
|
not modelled |
78.1 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 54 | d1rqba2 |
|
not modelled |
77.4 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:HMGL-like |
|
|
| 55 | c3qz6A_ |
|
not modelled |
77.4 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:hpch/hpai aldolase;
PDBTitle: the crystal structure of hpch/hpai aldolase from desulfitobacterium2 hafniense dcb-2
|
|
|
| 56 | d1ujqa_ |
|
not modelled |
76.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 57 | c5zjnB_ |
|
not modelled |
76.5 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:putative n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate-2-epimerase from vibrio2 cholerae with n-acetylmannosamine-6-phosphate
|
|
|
| 58 | d1qopa_ |
|
not modelled |
76.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 59 | c4uxdC_ |
|
not modelled |
76.0 |
9 |
PDB header:lyase
Chain: C: PDB Molecule:2-dehydro-3-deoxy-d-gluconate/2-dehydro-3-deoxy-
PDBTitle: 2-keto 3-deoxygluconate aldolase from picrophilus torridus
|
|
|
| 60 | c1zh8B_ |
|
not modelled |
75.1 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase;
PDBTitle: crystal structure of oxidoreductase (tm0312) from thermotoga maritima2 at 2.50 a resolution
|
|
|
| 61 | d1pkla2 |
|
not modelled |
74.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 62 | c2gzmB_ |
|
not modelled |
74.2 |
13 |
PDB header:isomerase
Chain: B: PDB Molecule:glutamate racemase;
PDBTitle: crystal structure of the glutamate racemase from bacillus anthracis
|
|
|
| 63 | c3ez4B_ |
|
not modelled |
74.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:3-methyl-2-oxobutanoate hydroxymethyltransferase;
PDBTitle: crystal structure of 3-methyl-2-oxobutanoate hydroxymethyltransferase2 from burkholderia pseudomallei
|
|
|
| 64 | c3s5oA_ |
|
not modelled |
74.0 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase, mitochondrial;
PDBTitle: crystal structure of human 4-hydroxy-2-oxoglutarate aldolase bound to2 pyruvate
|
|
|
| 65 | d1xlma_ |
|
not modelled |
73.4 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 66 | c5ks8F_ |
|
not modelled |
71.9 |
14 |
PDB header:ligase
Chain: F: PDB Molecule:pyruvate carboxylase subunit beta;
PDBTitle: crystal structure of two-subunit pyruvate carboxylase from2 methylobacillus flagellatus
|
|
|
| 67 | d1ur4a_ |
|
not modelled |
71.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 68 | c3pg8B_ |
|
not modelled |
71.6 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:phospho-2-dehydro-3-deoxyheptonate aldolase;
PDBTitle: truncated form of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase2 from thermotoga maritima
|
|
|
| 69 | c2y0fD_ |
|
not modelled |
71.6 |
21 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
|
|
|
| 70 | d2a6na1 |
|
not modelled |
71.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 71 | c2qiwA_ |
|
not modelled |
69.8 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 72 | d1pv8a_ |
|
not modelled |
69.3 |
42 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
|
|
| 73 | c6daoB_ |
|
not modelled |
69.1 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;
PDBTitle: nahe wt selenomethionine
|
|
|
| 74 | c4fxjB_ |
|
not modelled |
68.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:pyruvate kinase isozymes m1/m2;
PDBTitle: structure of m2 pyruvate kinase in complex with phenylalanine
|
|
|
| 75 | c3gr7A_ |
|
not modelled |
68.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: structure of oye from geobacillus kaustophilus, hexagonal crystal form
|
|
|
| 76 | c3tsmB_ |
|
not modelled |
67.8 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of indole-3-glycerol phosphate synthase from2 brucella melitensis
|
|
|
| 77 | c5ud6B_ |
|
not modelled |
67.5 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
| 78 | c5d88A_ |
|
not modelled |
67.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:predicted protease of the collagenase family;
PDBTitle: the structure of the u32 peptidase mk0906
|
|
|
| 79 | c3i4eA_ |
|
not modelled |
66.4 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:isocitrate lyase;
PDBTitle: crystal structure of isocitrate lyase from burkholderia2 pseudomallei
|
|
|
| 80 | d1muwa_ |
|
not modelled |
65.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 81 | c1jpkA_ |
|
not modelled |
65.5 |
20 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: gly156asp mutant of human urod, human uroporphyrinogen iii2 decarboxylase
|
|
|
| 82 | d1r3sa_ |
|
not modelled |
65.2 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:UROD/MetE-like
Family:Uroporphyrinogen decarboxylase, UROD |
|
|
| 83 | c3nvtA_ |
|
not modelled |
65.0 |
16 |
PDB header:transferase/isomerase
Chain: A: PDB Molecule:3-deoxy-d-arabino-heptulosonate 7-phosphate synthase;
PDBTitle: 1.95 angstrom crystal structure of a bifunctional 3-deoxy-7-2 phosphoheptulonate synthase/chorismate mutase (aroa) from listeria3 monocytogenes egd-e
|
|
|
| 84 | c5x8oA_ |
|
not modelled |
64.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: crystal structure of gmp reductase from trypanosoma brucei with2 guanosine 5'-triphosphate
|
|
|
| 85 | c4ph6A_ |
|
not modelled |
64.2 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: structure of 3-dehydroquinate dehydratase from enterococcus faecalis
|
|
|
| 86 | c5lzlH_ |
|
not modelled |
64.0 |
38 |
PDB header:lyase
Chain: H: PDB Molecule:delta-aminolevulinic acid dehydratase;
PDBTitle: pyrobaculum calidifontis 5-aminolaevulinic acid dehydratase
|
|
|
| 87 | c3noyA_ |
|
not modelled |
63.9 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
|
|
| 88 | c2r94B_ |
|
not modelled |
63.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:2-keto-3-deoxy-(6-phospho-)gluconate aldolase;
PDBTitle: crystal structure of kd(p)ga from t.tenax
|
|
|
| 89 | d2c1ha1 |
|
not modelled |
63.3 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
|
|
| 90 | d1o5ka_ |
|
not modelled |
63.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 91 | c4lrtC_ |
|
not modelled |
63.2 |
17 |
PDB header:lyase/oxidoreductase
Chain: C: PDB Molecule:4-hydroxy-2-oxovalerate aldolase;
PDBTitle: crystal and solution structures of the bifunctional enzyme2 (aldolase/aldehyde dehydrogenase) from thermomonospora curvata,3 reveal a cofactor-binding domain motion during nad+ and coa4 accommodation whithin the shared cofactor-binding site
|
|
|
| 92 | c2yw3E_ |
|
not modelled |
63.2 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:4-hydroxy-2-oxoglutarate aldolase/2-deydro-3-
PDBTitle: crystal structure analysis of the 4-hydroxy-2-oxoglutarate aldolase/2-2 deydro-3-deoxyphosphogluconate aldolase from tthb1
|
|
|
| 93 | d1d3ga_ |
|
not modelled |
63.1 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 94 | c6oviA_ |
|
not modelled |
62.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:keto-deoxy-phosphogluconate aldolase;
PDBTitle: crystal structure of kdpg aldolase from legionella pneumophila with2 pyruvate captured at low ph as a covalent carbinolamine intermediate
|
|
|
| 95 | d1ps9a1 |
|
not modelled |
62.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 96 | c1vs1B_ |
|
not modelled |
62.8 |
24 |
PDB header:transferase
Chain: B: PDB Molecule:3-deoxy-7-phosphoheptulonate synthase;
PDBTitle: crystal structure of 3-deoxy-d-arabino-heptulosonate-7-2 phosphate synthase (dahp synthase) from aeropyrum pernix3 in complex with mn2+ and pep
|
|
|
| 97 | d1o66a_ |
|
not modelled |
62.2 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 98 | c4b5sB_ |
|
not modelled |
62.2 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:4-hydroxy-2-oxo-heptane-1,7-dioate aldolase;
PDBTitle: crystal structures of divalent metal dependent pyruvate aldolase,2 hpai, in complex with pyruvate
|
|
|
| 99 | c3lciA_ |
|
not modelled |
61.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:n-acetylneuraminate lyase;
PDBTitle: the d-sialic acid aldolase mutant v251w
|
|
|
| 100 | c3navB_ |
|
not modelled |
61.9 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
|
|
|
| 101 | d2a0ma1 |
|
not modelled |
61.6 |
12 |
Fold:Arginase/deacetylase
Superfamily:Arginase/deacetylase
Family:Arginase-like amidino hydrolases |
|
|
| 102 | d1w3ia_ |
|
not modelled |
61.5 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 103 | c4ay8B_ |
|
not modelled |
60.7 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:methylcobalamin\: coenzyme m methyltransferase;
PDBTitle: semet-derivative of a methyltransferase from m. mazei
|
|
|
| 104 | d1ajza_ |
|
not modelled |
60.6 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Dihydropteroate synthetase-like
Family:Dihydropteroate synthetase |
|
|
| 105 | d1rvga_ |
|
not modelled |
60.2 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class II FBP aldolase |
|
|
| 106 | c5kzmA_ |
|
not modelled |
59.9 |
9 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-beta chain complex from2 francisella tularensis
|
|
|
| 107 | d1dvja_ |
|
not modelled |
59.4 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Decarboxylase |
|
|
| 108 | c2c3zA_ |
|
not modelled |
59.4 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:indole-3-glycerol phosphate synthase;
PDBTitle: crystal structure of a truncated variant of indole-3-2 glycerol phosphate synthase from sulfolobus solfataricus
|
|
|
| 109 | c1jvnB_ |
|
not modelled |
58.8 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:bifunctional histidine biosynthesis protein hishf;
PDBTitle: crystal structure of imidazole glycerol phosphate synthase: a tunnel2 through a (beta/alpha)8 barrel joins two active sites
|
|
|
| 110 | c2fglA_ |
|
not modelled |
58.8 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:alkaline thermostable endoxylanase;
PDBTitle: an alkali thermostable f/10 xylanase from alkalophilic bacillus sp.2 ng-27
|
|
|
| 111 | d2glka1 |
|
not modelled |
58.7 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Xylose isomerase |
|
|
| 112 | c2vwtA_ |
|
not modelled |
58.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:yfau, 2-keto-3-deoxy sugar aldolase;
PDBTitle: crystal structure of yfau, a metal ion dependent class ii2 aldolase from escherichia coli k12 - mg-pyruvate product3 complex
|
|
|
| 113 | c1zuwA_ |
|
not modelled |
57.5 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase 1;
PDBTitle: crystal structure of b.subtilis glutamate racemase (race) with d-glu
|
|
|
| 114 | c3l2iB_ |
|
not modelled |
57.3 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
|
|
|
| 115 | c2r8wB_ |
|
not modelled |
56.5 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
|
|
| 116 | c2ejaB_ |
|
not modelled |
56.4 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen decarboxylase;
PDBTitle: crystal structure of uroporphyrinogen decarboxylase from2 aquifex aeolicus
|
|
|
| 117 | c5uncB_ |
|
not modelled |
56.1 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoenolpyruvate phosphomutase;
PDBTitle: the crystal structure of phosphoenolpyruvate phosphomutase from2 streptomyces platensis subsp. rosaceus
|
|
|
| 118 | c6b8sB_ |
|
not modelled |
55.9 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dihydroorotate dehydrogenase (quinone);
PDBTitle: crystal structure of dihydroorotate dehydrogenase from helicobacter2 pylori with bound fmn
|
|
|
| 119 | c4gmfD_ |
|
not modelled |
55.8 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:yersiniabactin biosynthetic protein ybtu;
PDBTitle: apo structure of a thiazolinyl imine reductase from yersinia2 enterocolitica (irp3)
|
|
|
| 120 | c2qygC_ |
|
not modelled |
54.1 |
19 |
PDB header:unknown function
Chain: C: PDB Molecule:ribulose bisphosphate carboxylase-like protein 2;
PDBTitle: crystal structure of a rubisco-like protein rlp2 from rhodopseudomonas2 palustris
|
|
|