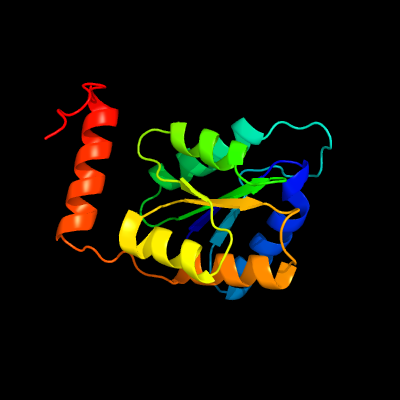
| 1 | d2vvpa1
|
|
 |
100.0 |
100 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
|
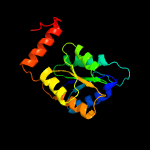
| 2 | c4lfnD_
|
|
 |
100.0 |
29 |
PDB header:isomerase
Chain: D: PDB Molecule:galactose-6-phosphate isomerase subunit b;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-ribulose
|
|
|
|
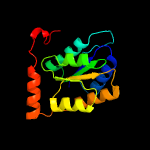
| 3 | c6mu0A_
|
|
 |
100.0 |
39 |
PDB header:isomerase
Chain: A: PDB Molecule:probable ribose-5-phosphate isomerase b;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b from mycoplasma2 genitalium with bound ribulose-5-phosphate
|
|
|
|
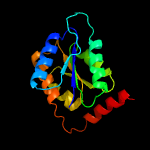
| 4 | c3he8A_
|
|
 |
100.0 |
37 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose-5-phosphate isomerase;
PDBTitle: structural study of clostridium thermocellum ribose-5-phosphate2 isomerase b
|
|
|
|
| 5 | c3qd5B_
|
|
 |
100.0 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:putative ribose-5-phosphate isomerase;
PDBTitle: crystal structure of a putative ribose-5-phosphate isomerase from2 coccidioides immitis solved by combined iodide ion sad and mr
|
|
|
|
| 6 | d1nn4a_
|
|
 |
100.0 |
32 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
|
| 7 | c6fxwA_
|
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:putative ribose 5-phosphate isomerase;
PDBTitle: structure of leishmania infantum type b ribose 5-phosphate isomerase
|
|
|
|
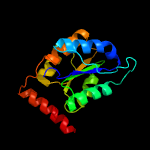
| 8 | c3m1pA_
|
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of ribose 5-phosphate isomerase type b from trypanosoma2 cruzi, soaked with allose-6-phosphate
|
|
|
|
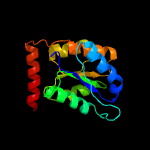
| 9 | c3k7pA_
|
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: structure of mutant of ribose 5-phosphate isomerase type b from2 trypanosoma cruzi.
|
|
|
|
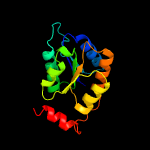
| 10 | d1o1xa_
|
|
 |
100.0 |
36 |
Fold:Ribose/Galactose isomerase RpiB/AlsB
Superfamily:Ribose/Galactose isomerase RpiB/AlsB
Family:Ribose/Galactose isomerase RpiB/AlsB |
|
|
|
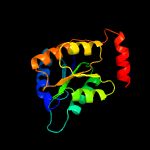
| 11 | c4lfmA_
|
|
 |
100.0 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:galactose-6-phosphate isomerase subunit a;
PDBTitle: crystal structure of d-galactose-6-phosphate isomerase in complex with2 d-psicose
|
|
|
|
| 12 | c4em8A_
|
|
 |
100.0 |
35 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase b;
PDBTitle: the structure of ribose 5-phosphate isomerase b from anaplasma2 phagocytophilum
|
|
|
|
| 13 | c6fxsA_
|
|
 |
100.0 |
36 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase, putative;
PDBTitle: structure of trypanosoma brucei type b ribose 5-phosphate isomerase
|
|
|
|
| 14 | c3s5pA_
|
|
 |
100.0 |
33 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase b rpib from giardia2 lamblia
|
|
|
|
| 15 | c5ifzA_
|
|
 |
100.0 |
34 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose 5-phosphate isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase from brucella2 melitensis 16m
|
|
|
|
| 16 | c3onoA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of ribose-5-phosphate isomerase lacab_rpib from2 vibrio parahaemolyticus
|
|
|
|
| 17 | c2ppwA_
|
|
 |
100.0 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: the crystal structure of uncharacterized ribose 5-phosphate isomerase2 rpib from streptococcus pneumoniae
|
|
|
|
| 18 | c3c5yD_
|
|
 |
100.0 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:ribose/galactose isomerase;
PDBTitle: crystal structure of a putative ribose 5-phosphate isomerase2 (saro_3514) from novosphingobium aromaticivorans dsm at 1.81 a3 resolution
|
|
|
|
| 19 | c2pjkA_
|
|
 |
94.6 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:178aa long hypothetical molybdenum cofactor
PDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
|
|
|
|
| 20 | d1y5ea1
|
|
 |
93.2 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
|
| 21 | d1mkza_ |
|
not modelled |
93.1 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 22 | c2xecD_ |
|
not modelled |
91.1 |
19 |
PDB header:isomerase
Chain: D: PDB Molecule:putative maleate isomerase;
PDBTitle: nocardia farcinica maleate cis-trans isomerase bound to2 tris
|
|
|
| 23 | c2is8A_ |
|
not modelled |
89.4 |
20 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
|
|
| 24 | d1v7ra_ |
|
not modelled |
88.1 |
11 |
Fold:Anticodon-binding domain-like
Superfamily:ITPase-like
Family:ITPase (Ham1) |
|
|
| 25 | c4fq5B_ |
|
not modelled |
87.0 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:maleate cis-trans isomerase;
PDBTitle: crystal structure of the maleate isomerase iso(c200a) from pseudomonas2 putida s16 with maleate
|
|
|
| 26 | c4xcwF_ |
|
not modelled |
87.0 |
18 |
PDB header:transferase
Chain: F: PDB Molecule:molybdopterin adenylyltransferase;
PDBTitle: crystal structure of molybdenum cofactor biosynthesis protein moga2 from helicobacter pylori str. j99
|
|
|
| 27 | c3rfqC_ |
|
not modelled |
84.6 |
22 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
|
|
| 28 | c6gyzB_ |
|
not modelled |
84.5 |
28 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: crystal structure of glmm from staphylococcus aureus
|
|
|
| 29 | d1p5dx1 |
|
not modelled |
83.5 |
33 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 30 | d1jlja_ |
|
not modelled |
83.2 |
12 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 31 | d1uuya_ |
|
not modelled |
82.4 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 32 | c3pdkB_ |
|
not modelled |
81.7 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: crystal structure of phosphoglucosamine mutase from b. anthracis
|
|
|
| 33 | c1tvmA_ |
|
not modelled |
80.2 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:pts system, galactitol-specific iib component;
PDBTitle: nmr structure of enzyme gatb of the galactitol-specific2 phosphoenolpyruvate-dependent phosphotransferase system
|
|
|
| 34 | c2rirA_ |
|
not modelled |
79.5 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
|
|
| 35 | c2f7lA_ |
|
not modelled |
79.4 |
23 |
PDB header:isomerase
Chain: A: PDB Molecule:455aa long hypothetical phospho-sugar mutase;
PDBTitle: crystal structure of sulfolobus tokodaii2 phosphomannomutase/phosphoglucomutase
|
|
|
| 36 | d2f7wa1 |
|
not modelled |
78.7 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 37 | d2g2ca1 |
|
not modelled |
78.6 |
20 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 38 | c3uw2A_ |
|
not modelled |
78.4 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase/phosphomannomutase family protein;
PDBTitle: x-ray crystal structure of phosphoglucomutase/phosphomannomutase2 family protein (bth_i1489)from burkholderia thailandensis
|
|
|
| 39 | c2qq1A_ |
|
not modelled |
78.4 |
21 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdenum cofactor biosynthesis mog;
PDBTitle: crystal structure of molybdenum cofactor biosynthesis2 (aq_061) other form from aquifex aeolicus vf5
|
|
|
| 40 | c3kbqA_ |
|
not modelled |
78.2 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein ta0487;
PDBTitle: the crystal structure of the protein cina with unknown function from2 thermoplasma acidophilum
|
|
|
| 41 | c4ix1B_ |
|
not modelled |
76.8 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of hypothetical protein opag_01669 from rhodococcus2 opacus pd630, target 016205
|
|
|
| 42 | d1di6a_ |
|
not modelled |
76.6 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 43 | d2ftsa3 |
|
not modelled |
76.3 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 44 | c1wqaB_ |
|
not modelled |
75.6 |
33 |
PDB header:isomerase
Chain: B: PDB Molecule:phospho-sugar mutase;
PDBTitle: crystal structure of pyrococcus horikoshii2 phosphomannomutase/phosphoglucomutase complexed with mg2+
|
|
|
| 45 | c5zi9B_ |
|
not modelled |
74.4 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of type-ii log from streptomyces coelicolor a3
|
|
|
| 46 | c2yxbA_ |
|
not modelled |
74.3 |
22 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
|
|
| 47 | c1vkrA_ |
|
not modelled |
73.4 |
29 |
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific pts system enzyme iiabc components;
PDBTitle: structure of iib domain of the mannitol-specific permease enzyme ii
|
|
|
| 48 | d1vkra_ |
|
not modelled |
73.4 |
29 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
|
|
| 49 | c1tuoA_ |
|
not modelled |
73.0 |
21 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative phosphomannomutase;
PDBTitle: crystal structure of putative phosphomannomutase from2 thermus thermophilus hb8
|
|
|
| 50 | d1uz5a3 |
|
not modelled |
71.5 |
15 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 51 | c1kgsA_ |
|
not modelled |
70.6 |
24 |
PDB header:dna binding protein
Chain: A: PDB Molecule:dna binding response regulator d;
PDBTitle: crystal structure at 1.50 a of an ompr/phob homolog from thermotoga2 maritima
|
|
|
| 52 | c3czcA_ |
|
not modelled |
70.6 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:rmpb;
PDBTitle: the crystal structure of a putative pts iib(ptxb) from streptococcus2 mutans
|
|
|
| 53 | d3bula2 |
|
not modelled |
68.7 |
22 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 54 | c2hunB_ |
|
not modelled |
68.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:336aa long hypothetical dtdp-glucose 4,6-dehydratase;
PDBTitle: crystal structure of hypothetical protein ph0414 from pyrococcus2 horikoshii ot3
|
|
|
| 55 | c3i3wB_ |
|
not modelled |
67.5 |
28 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucosamine mutase;
PDBTitle: structure of a phosphoglucosamine mutase from francisella tularensis
|
|
|
| 56 | c3vpbC_ |
|
not modelled |
66.2 |
19 |
PDB header:ligase
Chain: C: PDB Molecule:putative acetylornithine deacetylase;
PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
|
|
|
| 57 | d1p5dx2 |
|
not modelled |
65.5 |
18 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 58 | c3c04A_ |
|
not modelled |
64.1 |
30 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphomannomutase/phosphoglucomutase;
PDBTitle: structure of the p368g mutant of pmm/pgm from p. aeruginosa
|
|
|
| 59 | d2nqra3 |
|
not modelled |
62.1 |
31 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
|
|
| 60 | c1k98A_ |
|
not modelled |
61.1 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
|
|
| 61 | c4leiB_ |
|
not modelled |
60.7 |
28 |
PDB header:transferase
Chain: B: PDB Molecule:ndp-forosamyltransferase;
PDBTitle: spinosyn forosaminyltransferase spnp
|
|
|
| 62 | c3ezxA_ |
|
not modelled |
59.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
|
|
| 63 | c3ptjA_ |
|
not modelled |
58.0 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:upf0603 protein at1g54780, chloroplastic;
PDBTitle: structural and functional analysis of arabidopsis thaliana thylakoid2 lumen protein attlp18.3
|
|
|
| 64 | c2nqqA_ |
|
not modelled |
58.0 |
31 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:molybdopterin biosynthesis protein moea;
PDBTitle: moea r137q
|
|
|
| 65 | c3tquD_ |
|
not modelled |
56.3 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:non-canonical purine ntp pyrophosphatase;
PDBTitle: structure of a ham1 protein from coxiella burnetii
|
|
|
| 66 | c5bmpA_ |
|
not modelled |
55.7 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase;
PDBTitle: crystal structure of phosphoglucomutase from xanthomonas citri2 complexed with glucose-1-phosphate
|
|
|
| 67 | c2g4rB_ |
|
not modelled |
55.3 |
22 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:molybdopterin biosynthesis mog protein;
PDBTitle: anomalous substructure of moga
|
|
|
| 68 | c2fuvB_ |
|
not modelled |
54.7 |
28 |
PDB header:isomerase
Chain: B: PDB Molecule:phosphoglucomutase;
PDBTitle: phosphoglucomutase from salmonella typhimurium.
|
|
|
| 69 | c1yr3A_ |
|
not modelled |
52.9 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:xanthosine phosphorylase;
PDBTitle: escherichia coli purine nucleoside phosphorylase ii, the product of2 the xapa gene
|
|
|
| 70 | c3bq9A_ |
|
not modelled |
51.9 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
|
|
| 71 | c3d4oA_ |
|
not modelled |
51.7 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase subunit a;
PDBTitle: crystal structure of dipicolinate synthase subunit a (np_243269.1)2 from bacillus halodurans at 2.10 a resolution
|
|
|
| 72 | c5g2rA_ |
|
not modelled |
50.9 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:molybdopterin biosynthesis protein cnx1;
PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana
|
|
|
| 73 | c3dz1A_ |
|
not modelled |
50.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
|
|
| 74 | c4r3uD_ |
|
not modelled |
50.2 |
27 |
PDB header:isomerase
Chain: D: PDB Molecule:2-hydroxyisobutyryl-coa mutase small subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
| 75 | c4zrmB_ |
|
not modelled |
49.7 |
26 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of udp-glucose 4-epimerase (tm0509) from2 hyperthermophilic eubacterium thermotoga maritima
|
|
|
| 76 | c4mgeB_ |
|
not modelled |
48.7 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:pts system, cellobiose-specific iib component;
PDBTitle: 1.85 angstrom resolution crystal structure of pts system cellobiose-2 specific transporter subunit iib from bacillus anthracis.
|
|
|
| 77 | c5yvmA_ |
|
not modelled |
48.6 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the archaeal halo-thermophilic red sea brine pool2 alcohol dehydrogenase adh/d1 bound to nzq
|
|
|
| 78 | c1bmtB_ |
|
not modelled |
48.5 |
22 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
|
|
| 79 | c6oecD_ |
|
not modelled |
48.4 |
11 |
PDB header:structural protein
Chain: D: PDB Molecule:response regulator/sensory box protein/ggdef domain
PDBTitle: yeast spc42 trimeric coiled-coil amino acids 181-211 fused to pdb:2 3h5i
|
|
|
| 80 | c2x4gA_ |
|
not modelled |
48.4 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar epimerase2 from pseudomonas aeruginosa
|
|
|
| 81 | d1dbwa_ |
|
not modelled |
48.3 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 82 | d2gaxa1 |
|
not modelled |
46.3 |
11 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
|
|
| 83 | d1ka9h_ |
|
not modelled |
46.1 |
25 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
|
|
| 84 | c5gqsA_ |
|
not modelled |
45.0 |
19 |
PDB header:transport protein
Chain: A: PDB Molecule:pts galactitol transporter subunit iib;
PDBTitle: nmr based solution structure of pts system, galactitol-specific iib2 component from methicillin resistant staphylococcus aureus
|
|
|
| 85 | c2dgdD_ |
|
not modelled |
43.5 |
9 |
PDB header:lyase
Chain: D: PDB Molecule:223aa long hypothetical arylmalonate decarboxylase;
PDBTitle: crystal structure of st0656, a function unknown protein from2 sulfolobus tokodaii
|
|
|
| 86 | d1weha_ |
|
not modelled |
42.9 |
13 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 87 | c4hjhA_ |
|
not modelled |
42.4 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphomannomutase;
PDBTitle: iodide sad phased crystal structure of a phosphoglucomutase from2 brucella melitensis complexed with glucose-6-phosphate
|
|
|
| 88 | c2fu3A_ |
|
not modelled |
41.7 |
18 |
PDB header:biosynthetic protein/structural protein
Chain: A: PDB Molecule:gephyrin;
PDBTitle: crystal structure of gephyrin e-domain
|
|
|
| 89 | d1fmfa_ |
|
not modelled |
41.5 |
24 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 90 | c5iejA_ |
|
not modelled |
41.5 |
15 |
PDB header:protein
Chain: A: PDB Molecule:sdrg;
PDBTitle: solution structure of the bef3-activated conformation of sdrg from2 pseudomonas melonis fr1
|
|
|
| 91 | c5wq3A_ |
|
not modelled |
40.8 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:cytokinin riboside 5'-monophosphate phosphoribohydrolase;
PDBTitle: crystal strcuture of type-ii log from corynebacterium glutamicum
|
|
|
| 92 | c5f64C_ |
|
not modelled |
40.1 |
17 |
PDB header:transcription regulator
Chain: C: PDB Molecule:positive transcription regulator evga;
PDBTitle: putative positive transcription regulator (sensor evgs) from shigella2 flexneri
|
|
|
| 93 | d1gy8a_ |
|
not modelled |
39.8 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 94 | c3h5dD_ |
|
not modelled |
39.7 |
15 |
PDB header:lyase
Chain: D: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: dihydrodipicolinate synthase from drug-resistant streptococcus2 pneumoniae
|
|
|
| 95 | c2jjmH_ |
|
not modelled |
39.7 |
13 |
PDB header:transferase
Chain: H: PDB Molecule:glycosyl transferase, group 1 family protein;
PDBTitle: crystal structure of a family gt4 glycosyltransferase from bacillus2 anthracis orf ba1558.
|
|
|
| 96 | c2iyaB_ |
|
not modelled |
39.6 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:oleandomycin glycosyltransferase;
PDBTitle: the crystal structure of macrolide glycosyltransferases: a blueprint2 for antibiotic engineering
|
|
|
| 97 | d1ws6a1 |
|
not modelled |
37.8 |
15 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:YhhF-like |
|
|
| 98 | c1rcuB_ |
|
not modelled |
36.4 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein vt76;
PDBTitle: x-ray structure of tm1055 northeast structural genomics2 consortium target vt76
|
|
|
| 99 | d1rcua_ |
|
not modelled |
36.4 |
11 |
Fold:MCP/YpsA-like
Superfamily:MCP/YpsA-like
Family:MoCo carrier protein-like |
|
|
| 100 | c1y80A_ |
|
not modelled |
35.8 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from moorella2 thermoacetica
|
|
|
| 101 | c2i2xD_ |
|
not modelled |
35.8 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
|
|
| 102 | c2q1wC_ |
|
not modelled |
35.7 |
26 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
|
|
| 103 | c4yn8A_ |
|
not modelled |
35.4 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:response regulator chra;
PDBTitle: crystal structure of response regulator chra in heme-sensing two2 component system
|
|
|
| 104 | c3qayC_ |
|
not modelled |
35.1 |
17 |
PDB header:lyase
Chain: C: PDB Molecule:endolysin;
PDBTitle: catalytic domain of cd27l endolysin targeting clostridia difficile
|
|
|
| 105 | d3pmga1 |
|
not modelled |
35.0 |
20 |
Fold:Phosphoglucomutase, first 3 domains
Superfamily:Phosphoglucomutase, first 3 domains
Family:Phosphoglucomutase, first 3 domains |
|
|
| 106 | c6d9tA_ |
|
not modelled |
34.7 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyl transferase;
PDBTitle: bsha from staphylococcus aureus complexed with udp
|
|
|
| 107 | d1iiba_ |
|
not modelled |
34.1 |
13 |
Fold:Phosphotyrosine protein phosphatases I-like
Superfamily:PTS system IIB component-like
Family:PTS system, Lactose/Cellobiose specific IIB subunit |
|
|
| 108 | c5t3uA_ |
|
not modelled |
32.9 |
6 |
PDB header:transport protein
Chain: A: PDB Molecule:pts system, iia component;
PDBTitle: crystal structure of the pts iia protein associated with the fucose2 utilization operon from streptococcus pneumoniae
|
|
|
| 109 | c4m6iA_ |
|
not modelled |
32.6 |
27 |
PDB header:hydrolase
Chain: A: PDB Molecule:peptidoglycan amidase rv3717;
PDBTitle: structure of the reduced, zn-bound form of mycobacterium tuberculosis2 peptidoglycan amidase rv3717
|
|
|
| 110 | c3c4vB_ |
|
not modelled |
31.9 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:predicted glycosyltransferases;
PDBTitle: structure of the retaining glycosyltransferase msha:the2 first step in mycothiol biosynthesis. organism:3 corynebacterium glutamicum : complex with udp and 1l-ins-1-4 p.
|
|
|
| 111 | c4lhbC_ |
|
not modelled |
31.4 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:molybdopterin adenylyltransferase;
PDBTitle: crystal structure of tungsten cofactor synthesizing protein moab from2 pyrococcus furiosus
|
|
|
| 112 | d1qe5a_ |
|
not modelled |
31.1 |
26 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
|
|
| 113 | d1fp1d2 |
|
not modelled |
30.7 |
23 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Plant O-methyltransferase, C-terminal domain |
|
|
| 114 | c4s05B_ |
|
not modelled |
30.6 |
25 |
PDB header:transcription/dna
Chain: B: PDB Molecule:dna-binding transcriptional regulator basr;
PDBTitle: crystal structure of klebsiella pneumoniae pmra in complex with pmra2 box dna
|
|
|
| 115 | c4qs9A_ |
|
not modelled |
30.1 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:hexokinase-1;
PDBTitle: arabidopsis hexokinase 1 (athxk1) mutant s177a structure in glucose-2 bound form
|
|
|
| 116 | c3whpA_ |
|
not modelled |
30.1 |
13 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of the c-terminal domain of themus thermophilus litr2 in complex with cobalamin
|
|
|
| 117 | c4o6mA_ |
|
not modelled |
30.1 |
28 |
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
|
|
|
| 118 | c2esrB_ |
|
not modelled |
30.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: conserved hypothetical protein- streptococcus pyogenes
|
|
|
| 119 | c1kfiA_ |
|
not modelled |
29.3 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:phosphoglucomutase 1;
PDBTitle: crystal structure of the exocytosis-sensitive2 phosphoprotein, pp63/parafusin (phosphoglucomutase) from3 paramecium
|
|
|
| 120 | c2pzlB_ |
|
not modelled |
29.2 |
43 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmg in2 complex with nad and udp
|
|
|